1I0R
 
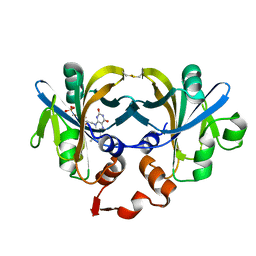 | | CRYSTAL STRUCTURE OF FERRIC REDUCTASE FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | CONSERVED HYPOTHETICAL PROTEIN, FLAVIN MONONUCLEOTIDE | | Authors: | Chiu, H.-J, Johnson, E, Schroder, I, Rees, D.C. | | Deposit date: | 2001-01-29 | | Release date: | 2001-05-02 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of a novel ferric reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus and its complex with NADP+.
Structure, 9, 2001
|
|
1L9J
 
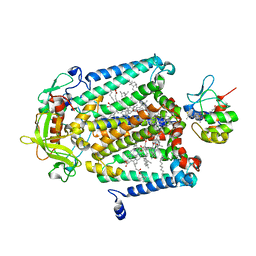 | | X-Ray Structure of the Cytochrome-c(2)-Photosynthetic Reaction Center Electron Transfer Complex from Rhodobacter sphaeroides in Type I Co-Crystals | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CHLORIDE ION, ... | | Authors: | Axelrod, H.L, Abresch, E.C, Okamura, M.Y, Yeh, A.P, Rees, D.C, Feher, G. | | Deposit date: | 2002-03-24 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | X-ray structure determination of the cytochrome c2: reaction center electron transfer complex from Rhodobacter sphaeroides.
J.Mol.Biol., 319, 2002
|
|
1L7V
 
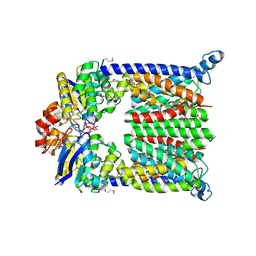 | | Bacterial ABC Transporter Involved in B12 Uptake | | Descriptor: | CYCLO-TETRAMETAVANADATE, VITAMIN B12 TRANSPORT SYSTEM PERMEASE PROTEIN BTUC, Vitamin B12 transport ATP-binding protein btuD | | Authors: | Locher, K.P, Lee, A.T, Rees, D.C. | | Deposit date: | 2002-03-18 | | Release date: | 2002-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism.
Science, 296, 2002
|
|
1L5H
 
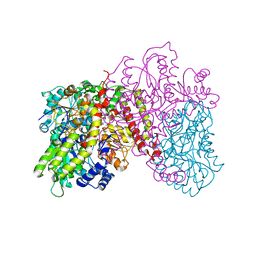 | | FeMo-cofactor Deficient Nitrogenase MoFe Protein | | Descriptor: | CALCIUM ION, FE(8)-S(7) CLUSTER, nitrogenase molybdenum-iron protein alpha chain, ... | | Authors: | Schmid, B, Ribbe, M.W, Einsle, O, Yoshida, M, Thomas, L.M, Dean, D.R, Rees, D.C, Burgess, B.K. | | Deposit date: | 2002-03-06 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a cofactor-deficient nitrogenase MoFe protein.
Science, 296, 2002
|
|
1L9B
 
 | | X-Ray Structure of the Cytochrome-c(2)-Photosynthetic Reaction Center Electron Transfer Complex from Rhodobacter sphaeroides in Type II Co-Crystals | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CHLORIDE ION, ... | | Authors: | Axelrod, H.L, Abresch, E.C, Okamura, M.Y, Yeh, A.P, Rees, D.C, Feher, G. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure determination of the cytochrome c2: reaction center electron transfer complex from Rhodobacter sphaeroides.
J.Mol.Biol., 319, 2002
|
|
1LRV
 
 | |
1M1Y
 
 | | Chemical Crosslink of Nitrogenase MoFe Protein and Fe Protein | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Schmid, B, Einsle, O, Chiu, H.J, Willing, A, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-20 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical and Structural Characterization of the Crosslinked Complex of Nitrogenase: Comparison to the ADP-AlF4- Stabilized Structure
Biochemistry, 41, 2002
|
|
1M1N
 
 | | Nitrogenase MoFe protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(7)-MO-S(9)-N CLUSTER, ... | | Authors: | Einsle, O, Tezcan, F.A, Andrade, S.L.A, Schmid, B, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-19 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Nitrogenase MoFe-protein at 1.16 A resolution: a central ligand in the FeMo-cofactor.
Science, 297, 2002
|
|
1M34
 
 | | Nitrogenase Complex From Azotobacter Vinelandii Stabilized By ADP-Tetrafluoroaluminate | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Schmid, B, Einsle, O, Chiu, H.-J, Willing, A, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-27 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Characterization of the Crosslinked Complex of Nitrogenase: Comparison to the ADP-AlF4 Stabilized structure
Biochemistry, 41, 2002
|
|
7JRF
 
 | | CO-CO-BOUND NITROGENASE MOFE-PROTEIN FROM A. VINELANDII | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, CARBON MONOXIDE, ... | | Authors: | Spatzal, T, Perez, K.A, Buscagan, T.M, Maggiolo, A.O, Rees, D.C. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural Characterization of Two CO Molecules Bound to the Nitrogenase Active Site.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7MC0
 
 | |
7MBZ
 
 | |
5CPA
 
 | |
6Q32
 
 | |
7AUV
 
 | | The structure of ERK2 in complex with dual inhibitor ASTX029 | | Descriptor: | (2~{R})-2-[5-[5-chloranyl-2-(oxan-4-ylamino)pyrimidin-4-yl]-3-oxidanylidene-1~{H}-isoindol-2-yl]-~{N}-[(1~{S})-1-(3-fluoranyl-5-methoxy-phenyl)-2-oxidanyl-ethyl]propanamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | O'Reilly, M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | ASTX029, a Novel Dual-mechanism ERK Inhibitor, Modulates Both the Phosphorylation and Catalytic Activity of ERK.
Mol.Cancer Ther., 20, 2021
|
|
3CPA
 
 | |
4AUA
 
 | | Liganded X-ray crystal structure of cyclin dependent kinase 6 (CDK6) | | Descriptor: | 1H-benzimidazol-2-yl(1H-pyrrol-2-yl)methanone, CYCLIN-DEPENDENT KINASE 6 | | Authors: | Cho, Y.S, Angove, H, Brain, C, Chen, C.H.T, Cheng, R, Chopra, R, Chung, K, Congreve, M, Dagostin, C, Davis, D, Feltell, R, Giraldes, J, Hiscock, S, Kim, S, Kovats, S, Lagu, B, Lewry, K, Loo, A, Lu, Y, Luzzio, M, Maniara, W, Mcmenamin, R, Mortenson, P, Benning, R, O'Reilly, M, Rees, D, Shen, J, Smith, T, Wang, Y, Williams, G, Woolford, A, Wrona, W, Xu, M, Yang, F, Howard, S. | | Deposit date: | 2012-05-15 | | Release date: | 2013-02-06 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
1AXM
 
 | | HEPARIN-LINKED BIOLOGICALLY-ACTIVE DIMER OF FIBROBLAST GROWTH FACTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | DiGabriele, A.D, Lax, I, Chen, D.I, Svahn, C.M, Jaye, M, Schlessinger, J, Hendrickson, W.A. | | Deposit date: | 1997-10-16 | | Release date: | 1998-04-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a heparin-linked biologically active dimer of fibroblast growth factor.
Nature, 393, 1998
|
|
7NR9
 
 | |
7NR8
 
 | | Discovery of ASTX029, a clinical candidate which modulates the phosphorylation and catalytic activity of ERK1/2 | | Descriptor: | (2~{R})-2-[5-[5-chloranyl-2-(oxan-4-ylamino)pyrimidin-4-yl]-3-oxidanylidene-1~{H}-isoindol-2-yl]-~{N}-[(1~{S})-1-[6-(4-methylpiperazin-1-yl)pyridin-2-yl]-2-oxidanyl-ethyl]propanamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | O'Reilly, M, Cleasby, A. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.627 Å) | | Cite: | Discovery of ASTX029, A Clinical Candidate Which Modulates the Phosphorylation and Catalytic Activity of ERK1/2.
J.Med.Chem., 64, 2021
|
|
7NR3
 
 | | Discovery of ASTX029, a clinical candidate which modulates the phosphorylation and catalytic activity of ERK1/2 | | Descriptor: | 6-[5-chloranyl-2-(oxan-4-ylamino)pyrimidin-4-yl]-2-[2-oxidanylidene-2-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)ethyl]-3~{H}-isoindol-1-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M. | | Deposit date: | 2021-03-02 | | Release date: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Discovery of ASTX029, A Clinical Candidate Which Modulates the Phosphorylation and Catalytic Activity of ERK1/2.
J.Med.Chem., 64, 2021
|
|
7NQQ
 
 | |
7NR5
 
 | | Discovery of ASTX029, a clinical candidate which modulates the phosphorylation and catalytic activity of ERK1/2 | | Descriptor: | (2~{R})-2-[5-[5-chloranyl-2-[(2-methyl-1,2,3-triazol-4-yl)amino]pyrimidin-4-yl]-3-oxidanylidene-1~{H}-isoindol-2-yl]-~{N}-[(1~{S})-1-(3-fluoranyl-5-methoxy-phenyl)-2-oxidanyl-ethyl]propanamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Discovery of ASTX029, A Clinical Candidate Which Modulates the Phosphorylation and Catalytic Activity of ERK1/2.
J.Med.Chem., 64, 2021
|
|
7NQW
 
 | | Discovery of ASTX029, a clinical candidate which modulates the phosphorylation and catalytic activity of ERK1/2 | | Descriptor: | 6-[5-chloranyl-2-(oxan-4-ylamino)pyrimidin-4-yl]-2-[2-oxidanylidene-2-(1,3,4,5-tetrahydro-2-benzazepin-2-yl)ethyl]-3~{H}-isoindol-1-one, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | O'Reilly, M. | | Deposit date: | 2021-03-02 | | Release date: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | Discovery of ASTX029, A Clinical Candidate Which Modulates the Phosphorylation and Catalytic Activity of ERK1/2.
J.Med.Chem., 64, 2021
|
|
3U7Q
 
 | |
