1LAG
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | HISTIDINE, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LST
 
 | |
8CHO
 
 | |
1W01
 
 | |
1W02
 
 | |
4UW2
 
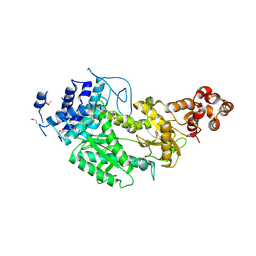 | | Crystal structure of Csm1 in T.onnurineus | | Descriptor: | CSM1 | | Authors: | Jung, T.Y, An, Y, Park, K.H, Lee, M.H, Oh, B.H, Woo, E.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-25 | | Last modified: | 2015-09-23 | | Method: | X-RAY DIFFRACTION (2.632 Å) | | Cite: | Crystal Structure of the Csm1 Subunit of the Csm Complex and its Single-Stranded DNA-Specific Nuclease Activity.
Structure, 23, 2015
|
|
2POO
 
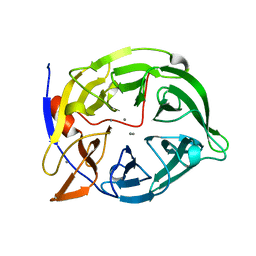 | | THERMOSTABLE PHYTASE IN FULLY CALCIUM LOADED STATE | | Descriptor: | CALCIUM ION, PROTEIN (PHYTASE) | | Authors: | Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-04-16 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
1CVM
 
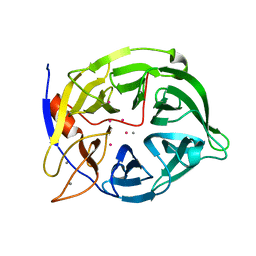 | | CADMIUM INHIBITED CRYSTAL STRUCTURE OF PHYTASE FROM BACILLUS AMYLOLIQUEFACIENS | | Descriptor: | CADMIUM ION, CALCIUM ION, PHYTASE | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-24 | | Release date: | 2000-02-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
7VT9
 
 | |
7VYT
 
 | | Crystal structure of human TIGIT(23-129) in complex with the scFv fragment of anti-TIGIT antibody MG1131 | | Descriptor: | CITRATE ANION, MG1131 heavy chain variable region, MG1131 light chain variable region, ... | | Authors: | Jeong, B.-S, Nam, H, Kim, M, Oh, B.-H. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and functional characterization of a monoclonal antibody blocking TIGIT.
Mabs, 14, 2022
|
|
7CBC
 
 | |
7DG5
 
 | |
4YJ4
 
 | | Crystal structure of Bcl-xL in complex with the BIM BH3 domain containing Ile155-to-Arg and Glu158-to-phosphoserine mutations | | Descriptor: | ACETATE ION, Bcl-2-like protein 1, BIM BH3 domain, ... | | Authors: | Ku, B, Ha, N.-C, Oh, B.-H. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-29 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conversion of cell-survival activity of Akt into apoptotic death of cancer cells by two mutations on the BIM BH3 domain.
Cell Death Dis, 6, 2015
|
|
1VZZ
 
 | |
7VYR
 
 | |
7OGT
 
 | | Folded elbow of cohesin | | Descriptor: | Structural maintenance of chromosomes protein 1, Structural maintenance of chromosomes protein 3 | | Authors: | Lee, B.-G, Gonzalez Llamazares, A, Collier, J, Patele, N.J, Nasmyth, K.A, Lowe, J. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Folding of cohesin's coiled coil is important for Scc2/4-induced association with chromosomes.
Elife, 10, 2021
|
|
4TSV
 
 | |
7XGE
 
 | |
7XGF
 
 | |
7XGG
 
 | |
6IWB
 
 | | Crystal structure of a computationally designed protein (LD3) in complex with BCL-2 | | Descriptor: | Apolipoprotein E, Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, SULFATE ION | | Authors: | Kim, S, Kwak, M.J, Oh, B.-H, Correia, B.E, Gainza, P. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A computationally designed chimeric antigen receptor provides a small-molecule safety switch for T-cell therapy.
Nat.Biotechnol., 38, 2020
|
|
6L7Q
 
 | |
1FXA
 
 | | CRYSTALLIZATION AND STRUCTURE DETERMINATION TO 2.5-ANGSTROMS RESOLUTION OF THE OXIDIZED [2FE-2S] FERREDOXIN ISOLATED FROM ANABAENA 7120 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, [2FE-2S] FERREDOXIN | | Authors: | Rypniewski, W.R, Breiter, D.R, Benning, M.M, Wesenberg, G, Oh, B.-H, Markley, J.L, Rayment, I, Holden, H.M. | | Deposit date: | 1991-01-09 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and structure determination to 2.5-A resolution of the oxidized [2Fe-2S] ferredoxin isolated from Anabaena 7120.
Biochemistry, 30, 1991
|
|
5TSW
 
 | |
2WMM
 
 | | Crystal structure of the hinge domain of MukB | | Descriptor: | Chromosome partition protein MukB, D-MALATE | | Authors: | Ku, B, Oh, B.-H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-12 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the MukB hinge domain with coiled-coil stretches and its functional implications.
Proteins, 78, 2010
|
|
