4MH7
 
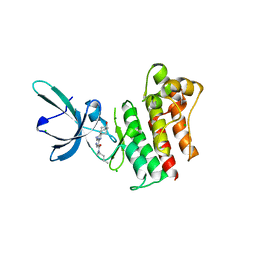 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1896 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-butyl-2-(butylamino)-4-[(trans-4-hydroxycyclohexyl)amino]-N-methylpyrimidine-5-carboxamide, ... | | Authors: | Zhang, W, McIver, A, Stashko, M.A, Deryckere, D, Branchford, B.R, Hunter, D, Kireev, D.B, Miley, M.J, Norris-Drouin, J, Stewart, W.M, Lee, M, Sather, S, Zhou, Y, DiPaola, J.A, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Mer specific tyrosine kinase inhibitors for the treatment and prevention of thrombosis.
J.Med.Chem., 56, 2013
|
|
3PJL
 
 | | The crystal structure of Tp34 bound to Co (II) ion at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, COBALT (II) ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Machius, M, Norgard, M.V. | | Deposit date: | 2010-11-10 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of the Tp34 lipoprotein from Treponema pallidum suggests a role in transition metal homeostasis
To be Published
|
|
3PJN
 
 | | The crystal structure of Tp34 bound to Zn(II) ion at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, SULFATE ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Machius, M, Norgard, M.V. | | Deposit date: | 2010-11-10 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of the Tp34 lipoprotein from Treponema pallidum suggests a role in transition metal homeostasis
To be Published
|
|
5VPA
 
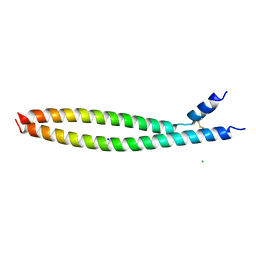 | | Transcription factor FosB/JunD bZIP domain | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPF
 
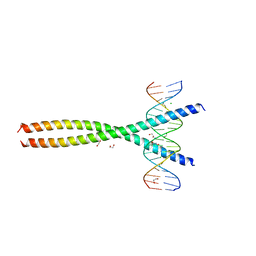 | | Transcription factor FosB/JunD bZIP domain in complex with cognate DNA, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*TP*CP*GP*GP*TP*GP*AP*CP*TP*CP*AP*CP*CP*GP*AP*CP*G)-3'), ... | | Authors: | Yin, Z, Rudenko, G, Machius, M. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPE
 
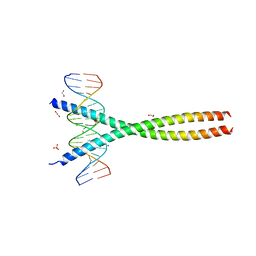 | | Transcription factor FosB/JunD bZIP domain in complex with cognate DNA, type-I crystal | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*TP*CP*GP*GP*TP*GP*AP*CP*TP*CP*AP*CP*CP*GP*AP*CP*G)-3'), ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
6UCM
 
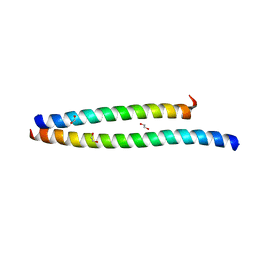 | | Transcription factor DeltaFosB bZIP domain self-assembly, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Protein fosB | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2019-09-16 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Self-assembly of the bZIP transcription factor Delta FosB.
Curr Res Struct Biol, 2, 2020
|
|
6UCI
 
 | | Transcription factor DeltaFosB bZIP domain self-assembly, oxidized form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein fosB | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2019-09-16 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Self-assembly of the bZIP transcription factor Delta FosB.
Curr Res Struct Biol, 2, 2020
|
|
6UCL
 
 | |
2IW5
 
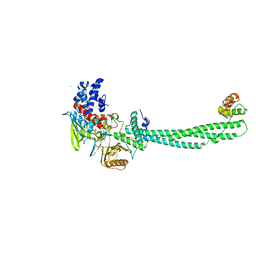 | | Structural Basis for CoREST-Dependent Demethylation of Nucleosomes by the Human LSD1 Histone Demethylase | | Descriptor: | AMMONIUM ION, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, M, Gocke, C.B, Luo, X, Borek, D, Tomchick, D.R, Machius, M, Otwinowski, Z, Yu, H. | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-09 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Basis for Corest-Dependent Demethylation of Nucleosomes by the Human Lsd1 Histone Demethylase
Mol.Cell, 23, 2006
|
|
3ONW
 
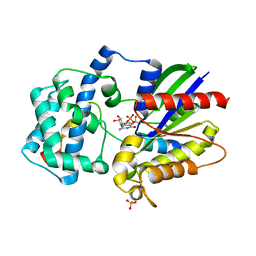 | | Structure of a G-alpha-i1 mutant with enhanced affinity for the RGS14 GoLoco motif. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, Regulator of G-protein signaling 14, ... | | Authors: | Bosch, D, Kimple, A.J, Sammond, D.W, Miley, M.J, Machius, M, Kuhlman, B, Willard, F.S, Siderovski, D.P. | | Deposit date: | 2010-08-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Determinants of Affinity Enhancement between GoLoco Motifs and G-Protein {alpha} Subunit Mutants.
J.Biol.Chem., 286, 2011
|
|
1X7Y
 
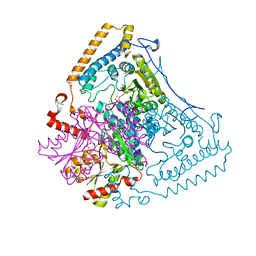 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1X7Z
 
 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1X80
 
 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1X7W
 
 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
5C7O
 
 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase holo form with NAD+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
1X7X
 
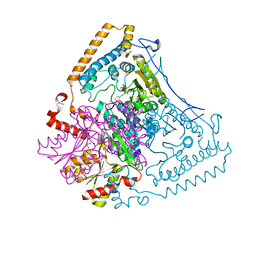 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
5C7L
 
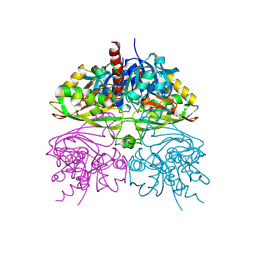 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase apo form | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
2VFX
 
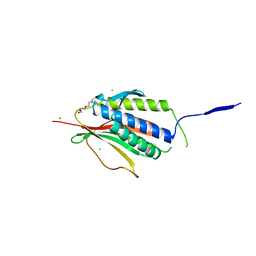 | | Structure of the Symmetric Mad2 Dimer | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, ... | | Authors: | Yang, M, Li, B, Liu, C.-J, Tomchick, D.R, Machius, M, Rizo, J, Yu, H, Luo, X. | | Deposit date: | 2007-11-05 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights Into MAD2 Regulation in the Spindle Checkpoint Revealed by the Crystal Structure of the Symmetric MAD2 Dimer.
Plos Biol., 6, 2008
|
|
1V11
 
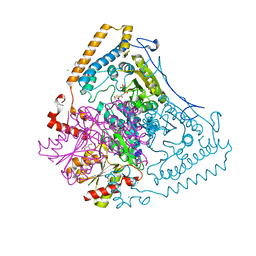 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
1V1M
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-20 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain Alpha-Keto Acid Decarboxylase/Dehydrogenase.
J.Biol.Chem., 279, 2004
|
|
1V1R
 
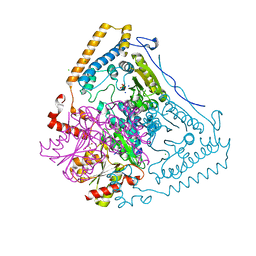 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, CHLORIDE ION, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-22 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
1V16
 
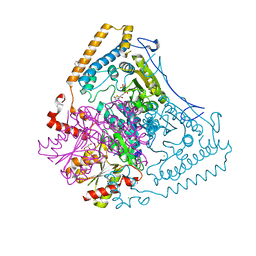 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-07 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
2FQY
 
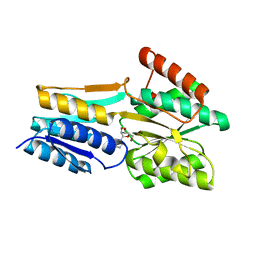 | | PnrA from Treponema pallidum complexed with adenosine. | | Descriptor: | ADENOSINE, Membrane lipoprotein tmpC | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Machius, M, Norgard, M.V. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The PnrA (Tp0319; TmpC) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC)-like operon in Treponema pallidum
J.Biol.Chem., 281, 2006
|
|
2FQW
 
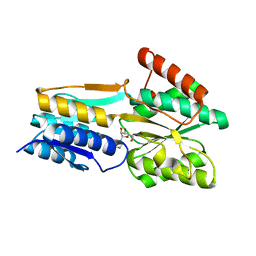 | | PnrA from Treponema pallidum as purified from E. coli (bound to inosine) | | Descriptor: | INOSINE, Membrane lipoprotein tmpC | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Machius, M, Norgard, M.V. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The PnrA (Tp0319; TmpC) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC)-like operon in Treponema pallidum
J.Biol.Chem., 281, 2006
|
|
