7RVO
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI13 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVT
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI20 | | Descriptor: | 3C-like proteinase, N~2~-[(2S)-2-{[(benzyloxy)carbonyl]amino}-2-cyclopropylacetyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVP
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI14 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-furan-2-yl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVZ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI26 | | Descriptor: | 3C-like proteinase, O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVW
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI23 | | Descriptor: | 3C-like proteinase, benzyl (1-{[(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamoyl}cyclopropyl)carbamate | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RW0
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI27 | | Descriptor: | 3C-like proteinase, N-{[(3-chlorophenyl)methoxy]carbonyl}-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVQ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI16 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVU
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI21 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-isovalyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
5X57
 
 | | Structure of GAR domain of ACF7 | | Descriptor: | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5, NICKEL (II) ION | | Authors: | Yang, F, Wang, T, Zhang, Y, Wu, X.Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | ACF7 regulates inflammatory colitis and intestinal wound response by orchestrating tight junction dynamics.
Nat Commun, 8, 2017
|
|
5Z9O
 
 | |
8H2T
 
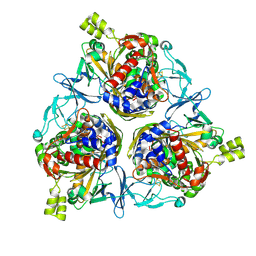 | | Cryo-EM structure of IadD/E dioxygenase bound with IAA | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Aromatic-ring-hydroxylating dioxygenase beta subunit, FE (III) ION, ... | | Authors: | Yu, G, Li, Z, Zhang, H. | | Deposit date: | 2022-10-07 | | Release date: | 2023-06-14 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural and biochemical characterization of the key components of an auxin degradation operon from the rhizosphere bacterium Variovorax.
Plos Biol., 21, 2023
|
|
7X5C
 
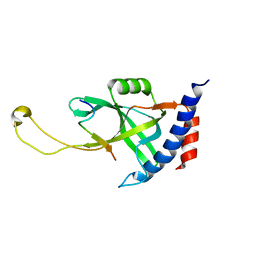 | | Solution structure of Tetrahymena p75OB1-p50PBM | | Descriptor: | Telomerase associated protein p50PBM, Telomerase-associated protein p75OB1 | | Authors: | Wu, B, Tang, T, Xue, H.J, Wu, J, Lei, M. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Association of the CST complex and p50 in Tetrahymena is crucial for telomere maintenance
Structure, 2022
|
|
7YW5
 
 | |
7XT2
 
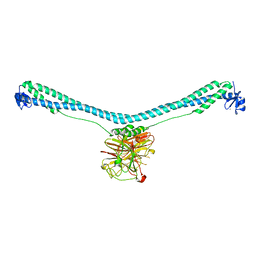 | | Crystal structure of TRIM72 | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Zhou, C, Ma, Y.M, Ding, L. | | Deposit date: | 2022-05-15 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for TRIM72 oligomerization during membrane damage repair.
Nat Commun, 14, 2023
|
|
4UEI
 
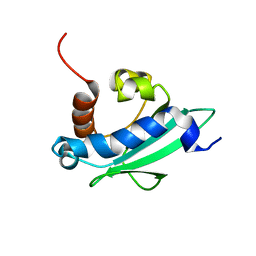 | | Solution structure of the sterol carrier protein domain 2 of Helicoverpa armigera | | Descriptor: | STEROL CARRIER PROTEIN 2/3-OXOACYL-COA THIOLASE | | Authors: | Liu, X, Ma, H, Yan, X, Hong, H, Peng, J, Peng, R. | | Deposit date: | 2014-12-18 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Function of Helicoverpa Armigera Sterol Carrier Protein-2, an Important Insecticidal Target from the Cotton Bollworm.
Sci.Rep., 5, 2015
|
|
8X8T
 
 | |
4XV2
 
 | |
4XV3
 
 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX7922 | | Descriptor: | N'-{3-[5-(2-aminopyrimidin-4-yl)-2-tert-butyl-1,3-thiazol-4-yl]-2-fluorophenyl}-N-ethyl-N-methylsulfuric diamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4XV9
 
 | | B-Raf Kinase domain in complex with PLX5568 | | Descriptor: | N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}-4-(trifluoromethyl)benzenesulfonamide, SULFATE ION, Serine/threonine-protein kinase B-raf | | Authors: | zhang, Y, zhang, c, wang, w. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4XV1
 
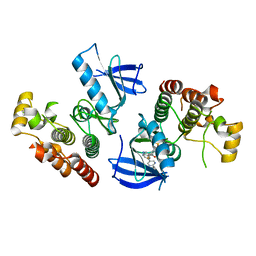 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX7904 | | Descriptor: | N'-(3-{[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)-N-ethyl-N-methylsulfuric diamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
6JCJ
 
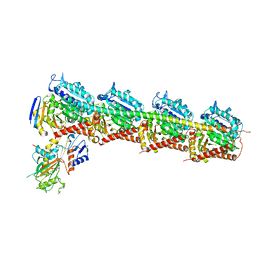 | | Structure of crolibulin in complex with tubulin | | Descriptor: | (4R)-2,7,8-triamino-4-(3-bromo-4,5-dimethoxyphenyl)-4H-1-benzopyran-3-carbonitrile, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z, Yang, J. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of crolibulin in complex with tubulin provides a rationale for drug design.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
7RPK
 
 | | Cryo-EM structure of murine Dispatched in complex with Sonic hedgehog | | Descriptor: | (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
4NIE
 
 | |
3JUG
 
 | |
7RPH
 
 | | Cryo-EM structure of murine Dispatched 'R' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
