5WX1
 
 | |
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | Descriptor: | Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5W
 
 | | Crystal structure of pseudorabies virus glycoprotein D | | Descriptor: | GD, Nectin-1 | | Authors: | Li, A, Lu, G, Qi, J, Wu, L, Tian, K, Luo, T, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2017-02-17 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pseudorabies virus glycoprotein D
To Be Published
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5V
 
 | | Crystal structure of pseudorabies virus glycoprotein D | | Descriptor: | GD | | Authors: | Li, A, Lu, G, Qi, J, Wu, L, Tian, K, Luo, T, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2017-02-17 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of pseudorabies virus glycoprotein D
To Be Published
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5Z38
 
 | | Crystal structure of CsrA bound to CesT | | Descriptor: | CesT protein, Truncated-CsrA, wild type CsrA | | Authors: | Ye, F, Yang, F, Yu, R, Lin, X, Qi, J, Chen, Z, Gao, G.F, Lu, G. | | Deposit date: | 2018-01-05 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Molecular basis of binding between the global post-transcriptional regulator CsrA and the T3SS chaperone CesT
Nat Commun, 9, 2018
|
|
5ZAN
 
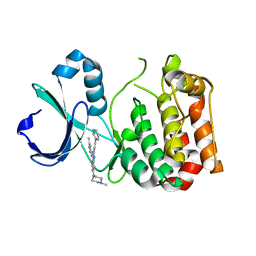 | |
6JIP
 
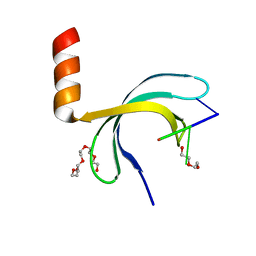 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), PENTAETHYLENE GLYCOL, SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6JIQ
 
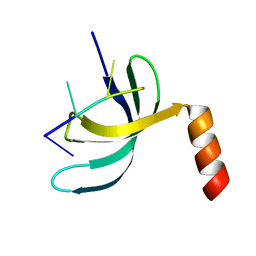 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6ADW
 
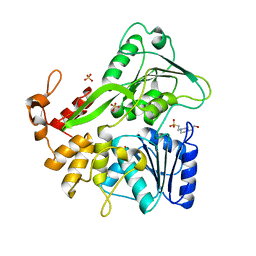 | | Crystal structure of the Zika virus NS3 helicase (apo form) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, Serine protease NS3 | | Authors: | Fang, J, Lu, G, Gong, P. | | Deposit date: | 2018-08-02 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Snapshots of the Zika Virus NS3 Helicase Help Visualize the Reactant Water Replenishment.
ACS Infect Dis, 5, 2019
|
|
6ADX
 
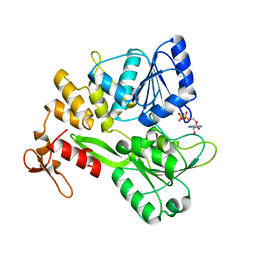 | | Crystal structure of the Zika virus NS3 helicase (ADP-Mn2+ complex, form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, Serine protease NS3 | | Authors: | Fang, J, Lu, G, Gong, P. | | Deposit date: | 2018-08-02 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystallographic Snapshots of the Zika Virus NS3 Helicase Help Visualize the Reactant Water Replenishment.
ACS Infect Dis, 5, 2019
|
|
6ADY
 
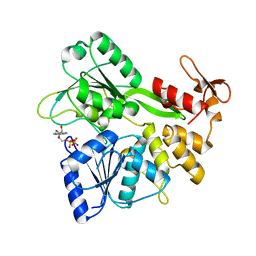 | | Crystal structure of the Zika virus NS3 helicase (ADP-Mn2+ complex, form 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, Serine protease NS3 | | Authors: | Fang, J, Lu, G, Gong, P. | | Deposit date: | 2018-08-02 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Snapshots of the Zika Virus NS3 Helicase Help Visualize the Reactant Water Replenishment.
ACS Infect Dis, 5, 2019
|
|
7XBK
 
 | | Structure and mechanism of a mitochondrial AAA+ disaggregase CLPB | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Isoform 2 of Caseinolytic peptidase B protein homolog, MAGNESIUM ION, ... | | Authors: | Wu, D, Liu, Y, Dai, Y, Wang, G, Lu, G, Chen, Y, Li, N, Lin, J, Gao, N. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
5F1B
 
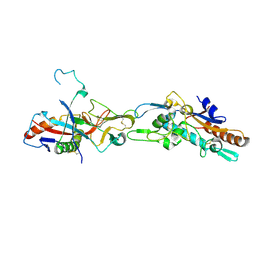 | | Structural basis of Ebola virus entry: viral glycoprotein bound to its endosomal receptor Niemann-Pick C1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GP1, GP2, ... | | Authors: | Wang, H, Shi, Y, Song, J, Qi, J, Lu, G, Yan, J, Gao, G.F. | | Deposit date: | 2015-11-30 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Cell, 164, 2016
|
|
5F18
 
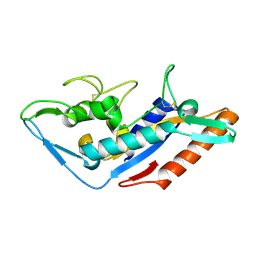 | | Structural basis of Ebola virus entry: viral glycoprotein bound to its endosomal receptor Niemann-Pick C1 | | Descriptor: | Niemann-Pick C1 protein | | Authors: | Wang, H, Shi, Y, Song, J, Qi, J, Lu, G, Yan, J, Gao, G.F. | | Deposit date: | 2015-11-30 | | Release date: | 2016-01-20 | | Last modified: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Cell, 164, 2016
|
|
4LMM
 
 | | Crystal structure of NHERF1 PDZ1 domain complexed with the CXCR2 C-terminal tail in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-07-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
4M7D
 
 | | Crystal structure of Lsm2-8 complex bound to the RNA fragment CGUUU | | Descriptor: | U6 snRNA, U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4M75
 
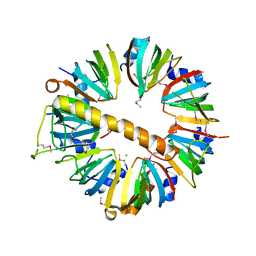 | | Crystal structure of Lsm1-7 complex | | Descriptor: | CHLORIDE ION, U6 snRNA-associated Sm-like protein Lsm1, U6 snRNA-associated Sm-like protein Lsm2, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4M77
 
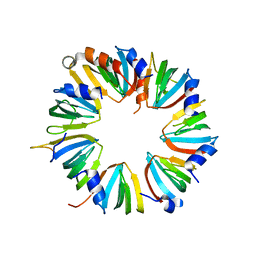 | | Crystal structure of Lsm2-8 complex, space group I212121 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.111 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4M78
 
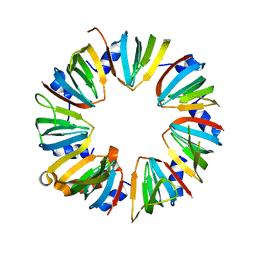 | | Crystal structure of Lsm2-8 complex, space group P21 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4M7A
 
 | | Crystal structure of Lsm2-8 complex bound to the 3' end sequence of U6 snRNA | | Descriptor: | U6 snRNA, U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
7XC5
 
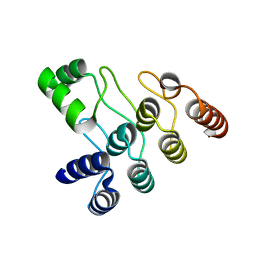 | | Crystal structure of the ANK domain of CLPB | | Descriptor: | Isoform 2 of Caseinolytic peptidase B protein homolog | | Authors: | Liu, Y, Wu, D, Lu, G, Gao, N, Lin, J. | | Deposit date: | 2022-03-23 | | Release date: | 2023-01-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
