6VDZ
 
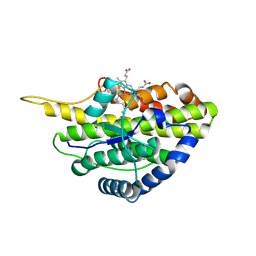 | | Crystal structure of reduced SfmD by soaking with sodium hydrosulfite | | Descriptor: | 3-methyl-L-tyrosine peroxygenase, HEME C | | Authors: | Shin, I, Liu, A. | | Deposit date: | 2019-12-27 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
6VE0
 
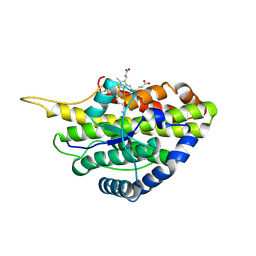 | | Crystal structure of reduced SfmD by soaking with sodium hydrosulfite | | Descriptor: | 3-methyl-L-tyrosine peroxygenase, HEME C | | Authors: | Shin, I, Liu, A. | | Deposit date: | 2019-12-27 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
8CZP
 
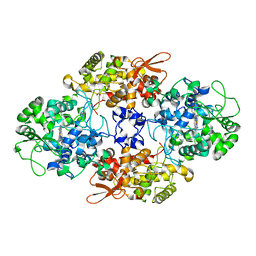 | |
5V27
 
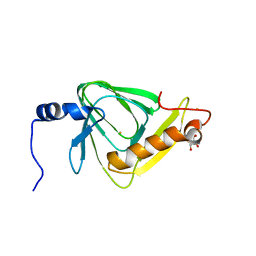 | | 2.35 angstrom crystal structure of P97V 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Dornevil, K, Liu, F, Liu, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | 2.35 angstrom crystal structure of P97V 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans
To Be Published
|
|
5V28
 
 | | 2.72 angstrom crystal structure of P97A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Dornevil, K, Liu, F, Liu, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | 2.72 angstrom crystal structure of P97A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans
To Be Published
|
|
5V26
 
 | |
1QJK
 
 | | Metallothionein MTA from sea urchin (alpha domain) | | Descriptor: | CADMIUM ION, METALLOTHIONEIN | | Authors: | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
1QJL
 
 | | METALLOTHIONEIN MTA FROM SEA URCHIN (BETA DOMAIN) | | Descriptor: | CADMIUM ION, METALLOTHIONEIN | | Authors: | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
4EWE
 
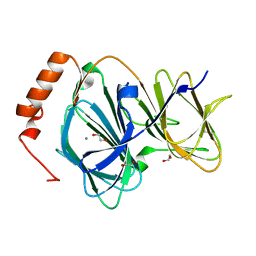 | | Study on structure and function relationships in human Pirin with Manganese ion | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Pirin | | Authors: | Liu, F, Rehmani, I, Fu, R, Esaka, S, Chen, L, Serrano, V, Liu, A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Pirin is an iron-dependent redox regulator of NF-kappa B.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EWD
 
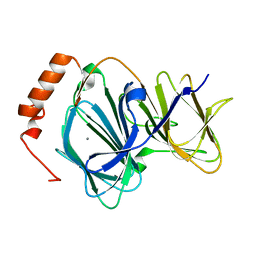 | | Study on structure and function relationships in human Pirin with Mn ion | | Descriptor: | MANGANESE (II) ION, Pirin | | Authors: | Liu, F, Rehmani, I, Chen, L, Fu, R, Serrano, V, Wilson, D.W, Liu, A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pirin is an iron-dependent redox regulator of NF-kappa B.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ERO
 
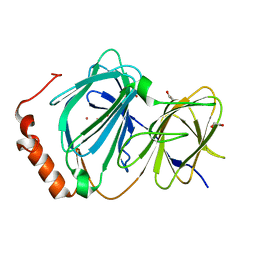 | | Study on structure and function relationships in human Pirin with Cobalt ion | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Pirin | | Authors: | Liu, F, Rehmani, I, Fu, R, Esaka, S, Chen, L, Serrano, V, Liu, A. | | Deposit date: | 2012-04-20 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Pirin is an iron-dependent redox regulator of NF-kappa B.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EWA
 
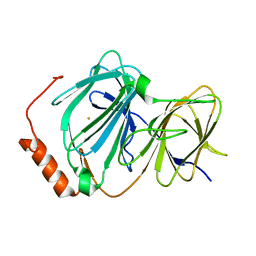 | | Study on structure and function relationships in human Pirin with Fe ion | | Descriptor: | FE (III) ION, Pirin | | Authors: | Liu, F, Rehmani, I, Chen, L, Fu, R, Serrano, V, Wilson, D.W, Liu, A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Pirin is an iron-dependent redox regulator of NF-kappa B.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GUL
 
 | | Study on structure and function relationships in human ferric Pirin | | Descriptor: | FE (III) ION, Pirin | | Authors: | Liu, F, Rehmani, I, Fu, R, Esaka, S, Chen, L, Serrano, V, Liu, A. | | Deposit date: | 2012-08-29 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pirin is an iron-dependent redox regulator of NF-kappa B.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6UPG
 
 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with cYF-4-OMe | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(4-methoxyphenyl)methyl]piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nguyen, R.C.D, Yang, Y, Liu, A. | | Deposit date: | 2019-10-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.393 Å) | | Cite: | Substrate-Assisted Hydroxylation and O-Demethylation in the Peroxidase-like Cytochrome P450 Enzyme CYP121
Acs Catalysis, 10, 2020
|
|
6UPI
 
 | | Crystal structure of Mycobacterium tuberculosis CYP121 bound with a hydroxylated intermediate of cYF-4-OMe | | Descriptor: | (3S,6S)-3-{[4-(hydroxymethoxy)phenyl]methyl}-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nguyen, R.C.D, Yang, Y, Liu, A. | | Deposit date: | 2019-10-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | Substrate-Assisted Hydroxylation and O-Demethylation in the Peroxidase-like Cytochrome P450 Enzyme CYP121
Acs Catalysis, 10, 2020
|
|
4HLT
 
 | | Crystal structure of ferric E32V Pirin | | Descriptor: | FE (II) ION, Pirin | | Authors: | Liu, F, Rehmani, I, Esaki, S, Fu, R, Chen, L, Serrano, V, Liu, A. | | Deposit date: | 2012-10-17 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pirin is an iron-dependent redox regulator of NF-kappa B.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HVR
 
 | | X-ray crystal structure of salicylic acid bound 3-hydroxyanthranilate-3,4-dioxygenase from cupriavidus metallidurans | | Descriptor: | 2-HYDROXYBENZOIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION | | Authors: | Liu, F, Chen, L, Liu, A. | | Deposit date: | 2012-11-06 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystal structure of salicylic acid bound 3-hydroxyanthranilate-3,4-dioxygenase from cupriavidus metallidurans
To be Published
|
|
4HSL
 
 | | 2.00 angstrom x-ray crystal structure of substrate-bound E110A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans | | Descriptor: | 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Liu, F, Chen, L, Liu, A. | | Deposit date: | 2012-10-30 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 angstrom x-ray crystal structure of substrate-bound
E110A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus
metallidurans
To be Published
|
|
4I3P
 
 | |
4IGM
 
 | | 2.39 Angstrom X-ray Crystal structure of human ACMSD | | Descriptor: | 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, ZINC ION | | Authors: | Liu, F, Liu, A. | | Deposit date: | 2012-12-17 | | Release date: | 2014-05-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | 2.39 Angstrom X-ray Crystal structure of human ACMSD
To be Published
|
|
4IGN
 
 | | 2.32 Angstrom X-ray Crystal structure of R47A mutant of human ACMSD | | Descriptor: | 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, ZINC ION | | Authors: | Liu, F, Liu, A. | | Deposit date: | 2012-12-17 | | Release date: | 2014-05-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | Human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD): A structural and mechanistic unveiling.
Proteins, 83, 2015
|
|
2HBX
 
 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
2HBV
 
 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
7KQ2
 
 | |
7KPZ
 
 | |
