7KJO
 
 | |
1PWV
 
 | |
1PWU
 
 | | Crystal Structure of Anthrax Lethal Factor complexed with (3-(N-hydroxycarboxamido)-2-isobutylpropanoyl-Trp-methylamide), a known small molecule inhibitor of matrix metalloproteases. | | Descriptor: | 3-(N-HYDROXYCARBOXAMIDO)-2-ISOBUTYLPROPANOYL-TRP-METHYLAMIDE, Lethal factor, ZINC ION | | Authors: | Wong, T.Y, Schwarzenbacher, R, Liddington, R.C. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1QG3
 
 | |
1QKR
 
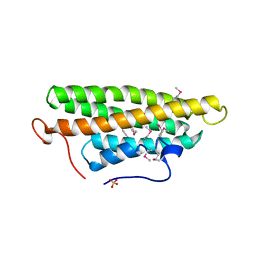 | | Crystal structure of the vinculin tail and a pathway for activation | | Descriptor: | SULFATE ION, VINCULIN | | Authors: | Bakolitsa, C, De Pereda, J.M, Bagshaw, C.R, Critchley, D.R, Liddington, R.C. | | Deposit date: | 1999-08-04 | | Release date: | 2000-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Vinculin Tail and a Pathway for Activation
Cell(Cambridge,Mass.), 99, 1999
|
|
7M1V
 
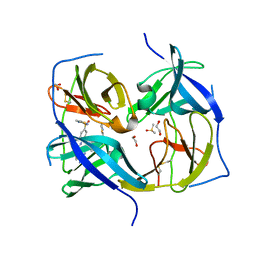 | | Structure of Zika virus NS2b-NS3 protease mutant binding the compound NSC86314 in the super-open conformation | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-{2,4-diamino-5-[2-(4-{[(2E)-1,3-thiazolidin-2-ylidene]sulfamoyl}phenyl)hydrazinyl]phenyl}hydrazinyl)-N-[(2S)-1,3-thiazolidin-2-yl]benzene-1-sulfonamide, CHLORIDE ION, ... | | Authors: | Aleshin, A.E, Shiryaev, S.A, Liddington, R.C. | | Deposit date: | 2021-03-15 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | To be provided
To Be Published
|
|
1RCW
 
 | |
4E18
 
 | | Alpha-E-catenin is an autoinhibited molecule that co-activates vinculin | | Descriptor: | Catenin alpha-1, Vinculin | | Authors: | Choi, H.-J, Pokutta, S, Cadwell, G.W, Bankston, L.A, Liddington, R.C, Weis, W.I. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Conformational plasticity of alpha-catenin revealed by binding interactions with vinculin
To be Published
|
|
4E17
 
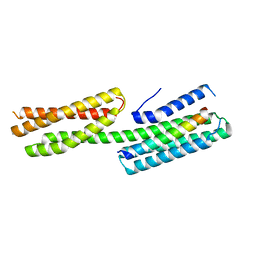 | | Alpha-E-catenin is an autoinhibited molecule that co-activates vinculin | | Descriptor: | Catenin alpha-1, Vinculin | | Authors: | Choi, H.-J, Pokutta, S, Cadwell, G.W, Bankston, L.A, Liddington, R.C, Weis, W.I. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Conformational plasticity of alpha-catenin revealed by binding interactions with vinculin
To be Published
|
|
1A4O
 
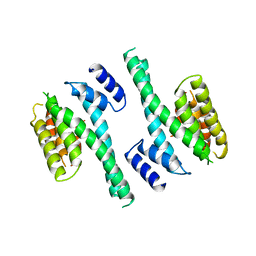 | | 14-3-3 PROTEIN ZETA ISOFORM | | Descriptor: | 14-3-3 PROTEIN ZETA | | Authors: | Liu, D, Bienkowska, J, Petosa, C, Collier, R.J, Fu, H, Liddington, R.C. | | Deposit date: | 1998-02-01 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the zeta isoform of the 14-3-3 protein.
Nature, 376, 1995
|
|
3RDR
 
 | | Structure of the catalytic domain of XlyA | | Descriptor: | CHLORIDE ION, N-acetylmuramoyl-L-alanine amidase XlyA, ZINC ION | | Authors: | Low, L.Y, Liddington, R.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins.
J.Biol.Chem., 286, 2011
|
|
3DYJ
 
 | |
1JLM
 
 | | I-DOMAIN FROM INTEGRIN CR3, MN2+ BOUND | | Descriptor: | INTEGRIN, MANGANESE (II) ION | | Authors: | Lee, J.-O, Liddington, R. | | Deposit date: | 1996-04-09 | | Release date: | 1997-01-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two conformations of the integrin A-domain (I-domain): a pathway for activation?
Structure, 3, 1995
|
|
1PDR
 
 | |
2H7E
 
 | |
2GJY
 
 | |
2H7D
 
 | |
1NIH
 
 | |
1IDO
 
 | | I-DOMAIN FROM INTEGRIN CR3, MG2+ BOUND | | Descriptor: | INTEGRIN, MAGNESIUM ION | | Authors: | Lee, J.-O, Liddington, R. | | Deposit date: | 1996-03-12 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18).
Cell(Cambridge,Mass.), 80, 1995
|
|
1V7P
 
 | | Structure of EMS16-alpha2-I domain complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EMS16 A chain, ... | | Authors: | Horii, K, Okuda, D, Morita, T, Mizuno, H. | | Deposit date: | 2003-12-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of EMS16 in complex with the integrin alpha2-I domain
J.Mol.Biol., 341, 2004
|
|
1BFE
 
 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 | | Descriptor: | PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
1BE9
 
 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 IN COMPLEX WITH A C-TERMINAL PEPTIDE DERIVED FROM CRIPT. | | Descriptor: | CRIPT, PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
1OTV
 
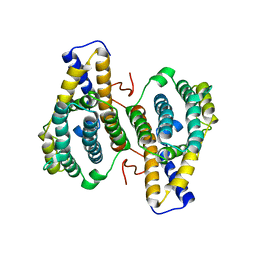 | | PqqC, Pyrroloquinolinquinone Synthase C | | Descriptor: | Coenzyme PQQ synthesis protein C | | Authors: | Magnusson, O.T, Toyama, H, Saeki, M, Rojas, A, Reed, J.C, Adachi, O, Klinman, J.P, SChwarzenbacher, R. | | Deposit date: | 2003-03-23 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Quinone Biogenesis: Structure and Mechanism of PqqC, the Final Catalyst in the Production of Pyrroloquinoline Quinone.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1COH
 
 | |
1G9O
 
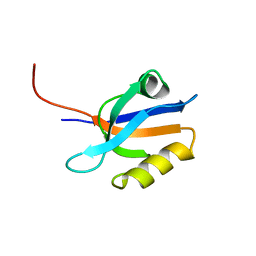 | | FIRST PDZ DOMAIN OF THE HUMAN NA+/H+ EXCHANGER REGULATORY FACTOR | | Descriptor: | NHE-RF | | Authors: | Karthikeyan, S, Leung, T, Birrane, G, Webster, G, Ladias, J.A.A. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the PDZ1 domain of human Na(+)/H(+) exchanger regulatory factor provides insights into the mechanism of carboxyl-terminal leucine recognition by class I PDZ domains.
J.Mol.Biol., 308, 2001
|
|
