1N6P
 
 | | Crystal Structure of Human Rab5a A30E mutant complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-5A | | Authors: | Zhu, G, Liu, J, Terzyan, S, Zhai, P, Li, G, Zhang, X.C. | | Deposit date: | 2002-11-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | High Resolution Crystal Structures of Human Rab5a and Five Mutants with Substitutions in the Catalytically Important Phosphate-Binding Loop
J.Biol.Chem., 278, 2003
|
|
1N6R
 
 | | Crystal Structure of Human Rab5a A30L mutant complex with GppNHp | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Zhu, G, Liu, J, Terzyan, S, Zhai, P, Li, G, Zhang, X.C. | | Deposit date: | 2002-11-11 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High Resolution Crystal Structures of Human Rab5a and Five Mutants with Substitutions in the Catalytically Important Phosphate-Binding Loop
J.Biol.Chem., 278, 2003
|
|
1R2Q
 
 | | Crystal Structure of Human Rab5a GTPase Domain at 1.05 A resolution | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Terzyan, S, Zhu, G, Li, G, Zhang, X.C. | | Deposit date: | 2003-09-29 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Refinement of the structure of human Rab5a GTPase domain at 1.05 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5DMM
 
 | | Crystal Structure of the Homocysteine Methyltransferase MmuM from Escherichia coli, Metallated form | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, BETA-MERCAPTOETHANOL, Homocysteine S-methyltransferase, ... | | Authors: | Li, K, Li, G, Bradbury, L.M.T, Andrew, H.D, Bruner, S.D. | | Deposit date: | 2015-09-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Crystal structure of the homocysteine methyltransferase MmuM from Escherichia coli.
Biochem.J., 473, 2016
|
|
5DML
 
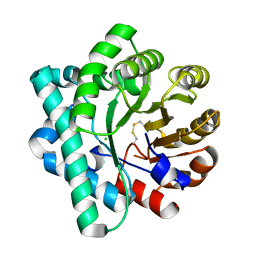 | | Crystal Structure of the Homocysteine Methyltransferase MmuM from Escherichia coli, Oxidized form | | Descriptor: | CHLORIDE ION, Homocysteine S-methyltransferase | | Authors: | Li, K, Li, G, Bradbury, L.M.T, Andrew, H.D, Bruner, S.D. | | Deposit date: | 2015-09-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Crystal structure of the homocysteine methyltransferase MmuM from Escherichia coli.
Biochem.J., 473, 2016
|
|
5DMN
 
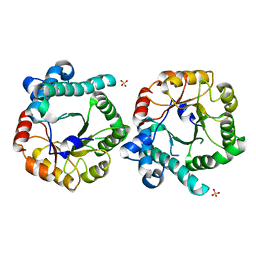 | | Crystal Structure of the Homocysteine Methyltransferase MmuM from Escherichia coli, Apo form | | Descriptor: | Homocysteine S-methyltransferase, SULFATE ION | | Authors: | Li, K, Li, G, Bradbury, L.M.T, Andrew, H.D, Bruner, S.D. | | Deposit date: | 2015-09-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | Crystal structure of the homocysteine methyltransferase MmuM from Escherichia coli.
Biochem.J., 473, 2016
|
|
3R45
 
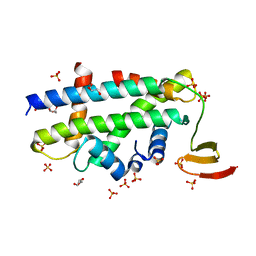 | | Structure of a CENP-A-Histone H4 Heterodimer in complex with chaperone HJURP | | Descriptor: | GLYCEROL, Histone H3-like centromeric protein A, Histone H4, ... | | Authors: | Hu, H, Liu, Y, Wang, M, Fang, J, Huang, H, Yang, N, Li, Y, Wang, J, Yao, X, Shi, Y, Li, G, Xu, R.M. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
Genes Dev., 25, 2011
|
|
2G9G
 
 | | Crystal structure of His-tagged mouse PNGase C-terminal domain | | Descriptor: | GLYCEROL, SULFATE ION, peptide N-glycanase | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2G9F
 
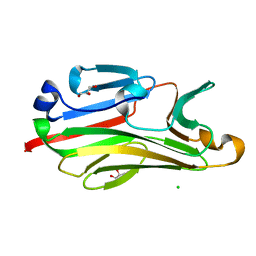 | | Crystal structure of intein-tagged mouse PNGase C-terminal domain | | Descriptor: | CHLORIDE ION, GLYCEROL, peptide N-glycanase | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HPL
 
 | | Crystal structure of the mouse p97/PNGase complex | | Descriptor: | C-terminal of mouse p97/VCP, PNGase | | Authors: | Zhao, G, Zhou, X, Wang, L, Li, G, Lennarz, W, Schindelin, H. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HPJ
 
 | | Crystal structure of the PUB domain of mouse PNGase | | Descriptor: | GLYCEROL, PNGase | | Authors: | Zhao, G, Zhou, X, Wang, L, Li, G, Lennarz, W, Schindelin, H. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2I74
 
 | | Crystal structure of mouse Peptide N-Glycanase C-terminal domain in complex with mannopentaose | | Descriptor: | ACETATE ION, GLYCEROL, PNGase, ... | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4N9B
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1-methyl-N-(pyridin-3-yl)-1H-pyrazole-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhai, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.859 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9D
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 4-({[(4-tert-butylphenyl)sulfonyl]amino}methyl)-N-(pyridin-3-yl)benzamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9C
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 5-nitro-1H-benzimidazole, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9E
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1-benzoylpiperidin-4-yl)methyl]-N-(pyridin-3-yl)-1H-benzimidazole-5-carboxamide, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2LGL
 
 | | NMR structure of the UHRF1 PHD domain | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
2LGG
 
 | | Structure of PHD domain of UHRF1 in complex with H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-26 | | Release date: | 2011-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
2LGK
 
 | | NMR Structure of UHRF1 PHD domains in a complex with histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
7CJL
 
 | | Metallo-Beta-Lactamase VIM-2 in complex with (S)-N-(3-(2H-tetrazol-5-yl)phenyl)-3-mercapto-2-methylpropanamide | | Descriptor: | (S)-N-(3-(2H-tetrazol-5-yl)phenyl)-3-mercapto-2-methylpropanamide, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Yan, Y.-H, Chen, J, Zhan, Z, Yu, Z.-J, Li, G, Li, G.-B, Guo, L, Wu, Y. | | Deposit date: | 2020-07-11 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Discovery of mercaptopropanamide-substituted aryl tetrazoles as new broad-spectrum metallo-beta-lactamase inhibitors.
Rsc Adv, 10, 2020
|
|
5ZRX
 
 | | Crystal Structure of EphA2/SHIP2 Complex | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2,Ephrin type-A receptor 2 | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
5ZRY
 
 | | Crystal Structure of EphA6/Odin Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Ankyrin repeat and SAM domain-containing protein 1A,Ephrin type-A receptor 6, ... | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
5ZRZ
 
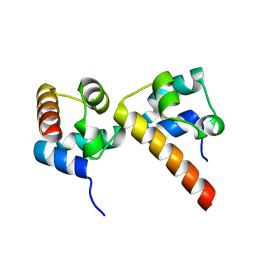 | | Crystal Structure of EphA5/SAMD5 Complex | | Descriptor: | Ephrin type-A receptor 5, Sterile alpha motif domain-containing protein 5 | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
6K61
 
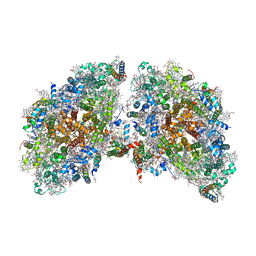 | | Cryo-EM structure of the tetrameric photosystem I from a heterocyst-forming cyanobacterium Anabaena sp. PCC7120 | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Zheng, L, Li, Y, Li, X, Zhong, Q, Li, N, Zhang, K, Zhang, Y, Chu, H, Ma, C, Li, G, Zhao, J, Gao, N. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural and functional insights into the tetrameric photosystem I from heterocyst-forming cyanobacteria.
Nat.Plants, 5, 2019
|
|
8G5S
 
 | | Crystal structure of apo TnmJ | | Descriptor: | TnmJ | | Authors: | Liu, Y.-C, Li, G, Gui, C, Shen, B. | | Deposit date: | 2023-02-14 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cofactorless oxygenases guide anthraquinone-fused enediyne biosynthesis.
Nat.Chem.Biol., 20, 2024
|
|
