7LYL
 
 | |
7LYK
 
 | |
7LYQ
 
 | |
7LYO
 
 | |
7LYP
 
 | |
7LYN
 
 | |
5YZV
 
 | | Biophysical and structural characterization of the thermostable WD40 domain of a prokaryotic protein, Thermomonospora curvata PkwA | | Descriptor: | Probable serine/threonine-protein kinase PkwA | | Authors: | Li, D.Y, Shen, C, Du, Y, Qiao, F.F, Kong, T, Yuan, L.R, Zhang, D.L, Wu, X.H, Wu, Y.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biophysical and structural characterization of the thermostable WD40 domain of a prokaryotic protein, Thermomonospora curvata PkwA
Sci Rep, 8, 2018
|
|
5WS2
 
 | | Crystal structure of mpy-RNase J (mutant S247A), an archaeal RNase J from Methanolobus psychrophilus R15, complex with RNA | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*A)-3'), Ribonuclease J, SULFATE ION, ... | | Authors: | Li, D.F, Feng, N. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | New molecular insights into an archaeal RNase J reveal a conserved processive exoribonucleolysis mechanism of the RNase J family
Mol. Microbiol., 106, 2017
|
|
8XON
 
 | |
8J5Z
 
 | | The cryo-EM structure of the TwOSC1 tetramer | | Descriptor: | Terpene cyclase/mutase family member, octyl beta-D-glucopyranoside | | Authors: | Ma, X, Yuru, T, Yunfeng, L, Jiang, T. | | Deposit date: | 2023-04-24 | | Release date: | 2023-11-01 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | Structural and Catalytic Insight into the Unique Pentacyclic Triterpene Synthase TwOSC.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8J8O
 
 | | Structure of Acb2 complexed with 2',3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, cGAMP, ... | | Authors: | Cao, X.L, Xiao, Y, Feng, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides.
Mol.Cell, 84, 2024
|
|
8JEY
 
 | |
8G6E
 
 | | Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304 | | Descriptor: | (7S,10S,13S)-N-cyclopentyl-10-[2-(morpholin-4-yl)ethyl]-9,12-dioxo-13-(2-oxopyrrolidin-1-yl)-2-oxa-8,11-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-7-carboxamide, Proteasome subunit alpha type, Proteasome subunit alpha type-1, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
8G6F
 
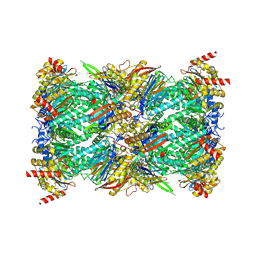 | | Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs | | Descriptor: | (2S)-N-[(E,2S)-1-(1H-indol-3-yl)-4-methylsulfonyl-but-3-en-2-yl]-2-[[(2S)-3-(1H-indol-3-yl)-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]-4-methyl-pentanamide, Proteasome endopeptidase complex, Proteasome subunit alpha type-1, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
8XN4
 
 | |
8XOO
 
 | |
8XOP
 
 | |
4XFV
 
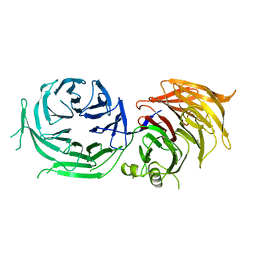 | | Crystal Structure of Elp2 | | Descriptor: | Elongator complex protein 2 | | Authors: | Lin, Z, Dong, C, Long, J, Shen, Y. | | Deposit date: | 2014-12-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The elp2 subunit is essential for elongator complex assembly and functional regulation
Structure, 23, 2015
|
|
6LLB
 
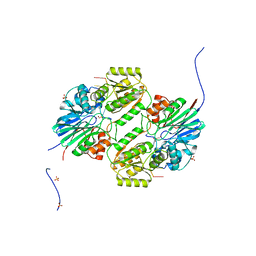 | | Crystal structure of mpy-RNase J (mutant S247A), an archaeal RNase J from Methanolobus psychrophilus R15, in complex with 6 nt RNA | | Descriptor: | MPY-RNase J, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, ... | | Authors: | Li, D.F, Hou, Y.J, Guo, L. | | Deposit date: | 2019-12-22 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A newly identified duplex RNA unwinding activity of archaeal RNase J depends on processive exoribonucleolysis coupled steric occlusion by its structural archaeal loops.
Rna Biol., 17, 2020
|
|
7WBL
 
 | | Cryo-EM structure of human ACE2 complexed with SARS-CoV-2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2021-12-17 | | Release date: | 2022-01-19 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2.
Cell, 185, 2022
|
|
7WBQ
 
 | |
7WBP
 
 | |
8IM6
 
 | | Crystal structure of HCoV 229E main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, Y.R, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of main proteases of HCoV-229E bound to inhibitor PF-07304814 and PF-07321332.
Biochem.Biophys.Res.Commun., 657, 2023
|
|
8QJJ
 
 | | CTE type II (tau intermediate amyloid) | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Scheres, S.H.W. | | Deposit date: | 2023-09-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
6IH0
 
 | | Aquifex aeolicus LpxC complex with ACHN-975 | | Descriptor: | N-[(2S)-3-azanyl-3-methyl-1-(oxidanylamino)-1-oxidanylidene-butan-2-yl]-4-[4-[(1R,2R)-2-(hydroxymethyl)cyclopropyl]buta -1,3-diynyl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION, ... | | Authors: | Li, D.Y, Fan, S, Jin, Y.Y, Zhang, C, Lv, G.X, Wu, G.T, Yang, Z.Y. | | Deposit date: | 2018-09-27 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Complex Structure of Protein AaLpxC from Aquifex aeolicus with ACHN-975 Molecule Suggests an Inhibitory Mechanism at Atomic-Level against Gram-Negative Bacteria
Molecules, 26, 2021
|
|
