6LKO
 
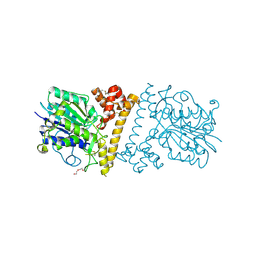 | |
5XC6
 
 | | Dengue Virus 4 NS3 Helicase in complex with SSRNA SLA12 | | Descriptor: | NS3 Helicase, PHOSPHATE ION, RNA (5'-R(*AP*GP*UP*UP*GP*UP*UP*AP*GP*UP*CP*U)-3') | | Authors: | Swarbrick, C.M.D, Basavannacharya, C, Chan, K.W.K, Chan, S.A, Singh, D, Wei, N, Phoo, W.W, Luo, D, Lescar, J, Vasudevan, S.G. | | Deposit date: | 2017-03-22 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication
Nucleic Acids Res., 45, 2017
|
|
5XC7
 
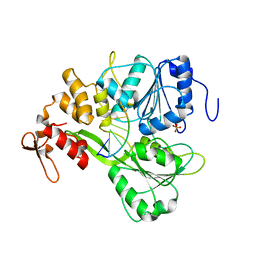
 | | Dengue Virus 4 NS3 Helicase D290A mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, NS3 Helicase | | Authors: | Swarbrick, C.M.D, Basavannacharya, C, Chan, K.W.K, Chan, S.A, Singh, D, Wei, N, Phoo, W.W, Luo, D, Lescar, J, Vasudevan, S.G. | | Deposit date: | 2017-03-22 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication
Nucleic Acids Res., 45, 2017
|
|
5XVU
 
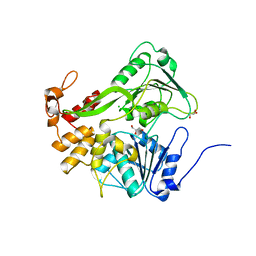
 | | Crystal structure of the protein kinase CK2 catalytic domain from Plasmodium falciparum bound to ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | El Sahili, A, Lescar, J, Ruiz-Carrillo, D, Lin, J.Q. | | Deposit date: | 2017-06-28 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The protein kinase CK2 catalytic domain from Plasmodium falciparum: crystal structure, tyrosine kinase activity and inhibition.
Sci Rep, 8, 2018
|
|
5ZHI
 
 | | Apo crystal structure of TrmD from Mycobacterium tuberculosis | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5ZHM
 
 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-({4-[(diethylamino)methyl]phenyl}methyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5ZHK
 
 | | Crystal structure of TrmD from Mycobacterium tuberculosis in complex with active-site inhibitor | | Descriptor: | N-[(4-{[cyclohexyl(ethyl)amino]methyl}phenyl)methyl]-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5ZHN
 
 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-({4-[(octylamino)methyl]phenyl}methyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5ZHJ
 
 | | Crystal structure of TrmD from Mycobacterium tuberculosis in complex with S-adenosyl homocysteine (SAH) | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5ZHL
 
 | | Crystal structure of TrmD from Mycobacterium tuberculosis in complex with active-site inhibitor | | Descriptor: | N-({4-[(octylamino)methyl]phenyl}methyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5ZBI
 
 | |
7VB4
 
 | |
7VW5
 
 | |
1URZ
 
 | |
5K5M
 
 | |
6S8C
 
 | |
2MAU
 
 | |
4ADG
 
 | | Crystal structure of the Rubella virus envelope Glycoprotein E1 in post-fusion form (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | DuBois, R.M, Vaney, M.C, Tortorici, M.A, Al Kurdi, R, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2011-12-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Functional and Evolutionary Insight from the Crystal Structure of Rubella Virus Protein E1.
Nature, 493, 2013
|
|
4ADI
 
 | | Crystal structure of the Rubella virus envelope glycoprotein E1 in post-fusion form (crystal form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | DuBois, R.M, Vaney, M.C, Tortorici, M.A, Al Kurdi, R, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2011-12-26 | | Release date: | 2013-01-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and Evolutionary Insight from the Crystal Structure of Rubella Virus Protein E1.
Nature, 493, 2013
|
|
4ADJ
 
 | | Crystal structure of the Rubella virus glycoprotein E1 in its post-fusion form crystallized in presence of 1mM of calcium acetate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | DuBois, R.M, Vaney, M.C, Tortorici, M.A, Al Kurdi, R, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2011-12-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Functional and Evolutionary Insight from the Crystal Structure of Rubella Virus Protein E1.
Nature, 493, 2013
|
|
4B3V
 
 | | Crystal structure of the Rubella virus glycoprotein E1 in its post-fusion form crystallized in presence of 20mM of Calcium Acetate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Vaney, M.C, DuBois, R.M, Tortorici, M.A, Rey, F.A. | | Deposit date: | 2012-07-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Functional and Evolutionary Insight from the Crystal Structure of Rubella Virus Protein E1.
Nature, 493, 2013
|
|
3PA7
 
 | | Crystal structure of FKBP from plasmodium vivax in complex with tetrapeptide ALPF | | Descriptor: | 4-mer Peptide ALPF, 70 kDa peptidylprolyl isomerase, putative | | Authors: | Balakrishna, A.M, Alag, R, Yoon, H.S. | | Deposit date: | 2010-10-18 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insights into substrate binding by PvFKBP35, a peptidylprolyl cis-trans isomerase from the human malarial parasite Plasmodium vivax
EUKARYOTIC CELL, 12, 2013
|
|
4ITZ
 
 | |
5E6X
 
 | |
5E6W
 
 | |
