2YNR
 
 | | mImp_alphadIBB_B54NLS | | Descriptor: | B54NLS, IMPORTIN SUBUNIT ALPHA | | Authors: | Chang, C.-W, Counago, R.L.M, Williams, S.J, Boden, M, Kobe, B. | | Deposit date: | 2012-10-18 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Rice Importin-Alpha and Structural Basis of its Interaction with Plant-Specific Nuclear Localization Signals.
Plant Cell, 24, 2012
|
|
5KU7
 
 | |
4UTO
 
 | | Crystal structure of pneumococcal surface antigen PsaA D280N in the Cd-bound, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
4UTP
 
 | | Crystal structure of pneumococcal surface antigen PsaA in the Cd- bound, closed state | | Descriptor: | CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
2Q2B
 
 | |
2QVT
 
 | | Structure of Melampsora lini avirulence protein, AvrL567-D | | Descriptor: | AvrL567-D | | Authors: | Guncar, G, Wang, C.I, Forwood, J.K, Teh, T, Catanzariti, A.M, Lawrence, G, Schirra, H.J, Anderson, P.A, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity.
Plant Cell, 19, 2007
|
|
2V1O
 
 | | Crystal structure of N-terminal domain of acyl-CoA thioesterase 7 | | Descriptor: | COENZYME A, CYTOSOLIC ACYL COENZYME A THIOESTER HYDROLASE | | Authors: | Forwood, J.K, Thakur, A.S, Guncar, G, Marfori, M, Mouradov, D, Meng, W.N, Robinson, J, Huber, T, Kellie, S, Martin, J.L, Hume, D.A, Kobe, B. | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Recruitment of Tandem Hotdog Domains in Acyl-Coa Thioesterase 7 and its Role in Inflammation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4ZM5
 
 | | Shigella flexneri lipopolysaccharide O-antigen chain-length regulator WzzBSF - A107P mutant | | Descriptor: | CHLORIDE ION, Chain length determinant protein, MAGNESIUM ION | | Authors: | Ericsson, D.J, Chang, C.-W, Lonhienne, T, Casey, L, Benning, F, Kobe, B, Tran, E.N.H, Morona, R. | | Deposit date: | 2015-05-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural and Biochemical Analysis of a Single Amino-Acid Mutant of WzzBSF That Alters Lipopolysaccharide O-Antigen Chain Length in Shigella flexneri.
Plos One, 10, 2015
|
|
4ZM1
 
 | | Shigella flexneri lipopolysaccharide O-antigen chain-length regulator WzzBSF - wild type | | Descriptor: | CITRIC ACID, Chain length determinant protein, MAGNESIUM ION | | Authors: | Ericsson, D.J, Chang, C.-W, Lonhienne, T, Casey, L, Benning, F, Kobe, B, Tran, E.N.H, Morona, R. | | Deposit date: | 2015-05-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Biochemical Analysis of a Single Amino-Acid Mutant of WzzBSF That Alters Lipopolysaccharide O-Antigen Chain Length in Shigella flexneri.
Plos One, 10, 2015
|
|
6CSL
 
 | | Pneumococcal PhtD protein 269-339 fragment with bound Zn(II) | | Descriptor: | Histidine triad protein D, ZINC ION | | Authors: | Luo, Z, Pederick, V.G, Paton, J.C, McDevitt, C.A, Kobe, B. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Structural characterisation of the HT3 motif of the polyhistidine triad protein D from Streptococcus pneumoniae.
FEBS Lett., 592, 2018
|
|
4DWN
 
 | | Crystal Structure of Human BinCARD CARD | | Descriptor: | Bcl10-interacting CARD protein, SULFATE ION | | Authors: | Chen, K.-E, Kobe, B, Martin, J.L. | | Deposit date: | 2012-02-26 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | The structure of the caspase recruitment domain of BinCARD reveals that all three cysteines can be oxidized.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FH0
 
 | | Crystal Structure of Human BinCARD CARD, double mutant F16M/L66M SeMet form | | Descriptor: | Bcl10-interacting CARD protein, SULFATE ION | | Authors: | Chen, K.-E, Kobe, B, Martin, J.L. | | Deposit date: | 2012-06-05 | | Release date: | 2013-02-06 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of the caspase recruitment domain of BinCARD reveals that all three cysteines can be oxidized.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4MZ5
 
 | | Structure of importin-alpha: dUTPase NLS complex | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial, Importin subunit alpha-1 | | Authors: | Marfori, M, Rona, G, Vertessy, B.G, Kobe, B. | | Deposit date: | 2013-09-29 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphorylation adjacent to the nuclear localization signal of human dUTPase abolishes nuclear import: structural and mechanistic insights.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4MZ6
 
 | | Structure of importin-alpha: dUTPase S11E NLS mutant complex | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial, Importin subunit alpha-1 | | Authors: | Marfori, M, Rona, G, Vertessy, B.G, Kobe, B. | | Deposit date: | 2013-09-29 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Phosphorylation adjacent to the nuclear localization signal of human dUTPase abolishes nuclear import: structural and mechanistic insights.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7UWG
 
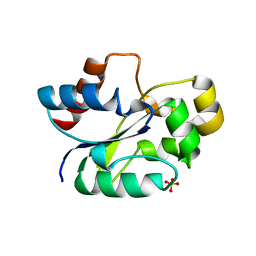 | | The crystal structure of the TIR domain-containing protein from Acinetobacter baumannii (AbTir) | | Descriptor: | HEXAETHYLENE GLYCOL, Molecular chaperone Tir, SULFATE ION | | Authors: | Manik, M.K, Nanson, J.D, Ve, T, Kobe, B. | | Deposit date: | 2022-05-03 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXU
 
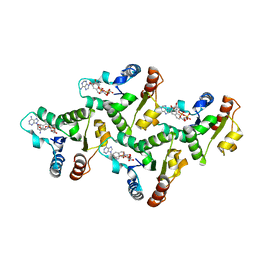 | | CryoEM structure of the TIR domain from AbTir in complex with 3AD | | Descriptor: | Molecular chaperone Tir, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Li, S, Nanson, J.D, Manik, M.K, Gu, W, Landsberg, M.J, Ve, T, Kobe, B. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
1Q1S
 
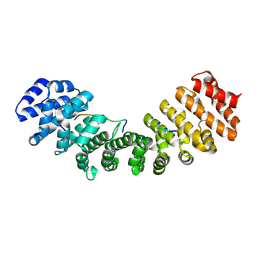 | | Mouse Importin alpha- phosphorylated SV40 CN peptide complex | | Descriptor: | Importin alpha-2 subunit, Large T antigen | | Authors: | Fontes, M.R.M, Teh, T, Toth, G, John, A, Pavo, I, Jans, D.A, Kobe, B. | | Deposit date: | 2003-07-22 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha
Biochem.J., 375, 2003
|
|
1Q1T
 
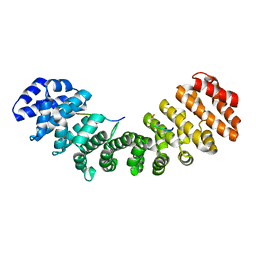 | | Mouse Importin alpha: non-phosphorylated SV40 CN peptide complex | | Descriptor: | Importin alpha-2 subunit, Large T antigen | | Authors: | Fontes, M.R.M, Teh, T, Toth, G, John, A, Pavo, I, Jans, D.A, Kobe, B. | | Deposit date: | 2003-07-22 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha
Biochem.J., 375, 2003
|
|
1WNH
 
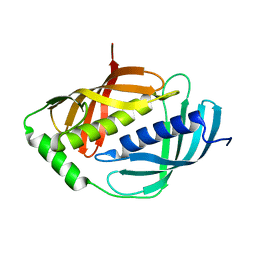 | | Crystal structure of mouse Latexin (tissue carboxypeptidase inhibitor) | | Descriptor: | Latexin | | Authors: | Aagaard, A, Listwan, P, Cowieson, N, Huber, T, Ravasi, T, Wells, C.A, Flanagan, J.U, Hume, D.A, Kobe, B, Martin, J.L. | | Deposit date: | 2004-08-04 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An Inflammatory Role for the Mammalian Carboxypeptidase Inhibitor Latexin: Relationship to Cystatins and the Tumor Suppressor TIG1
Structure, 13, 2005
|
|
7SHJ
 
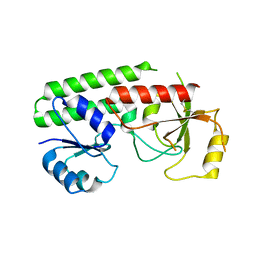 | |
7SZL
 
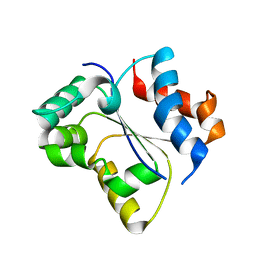 | | Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1R10 (IL-1RAPL2) | | Descriptor: | X-linked interleukin-1 receptor accessory protein-like 2 | | Authors: | Nimma, S, Gu, W, Manik, M.K, Ve, T, Nanson, J.D, Kobe, B. | | Deposit date: | 2021-11-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Toll/interleukin-1 receptor (TIR) domain of IL-1R10 provides structural insights into TIR domain signalling.
Febs Lett., 596, 2022
|
|
7T4Z
 
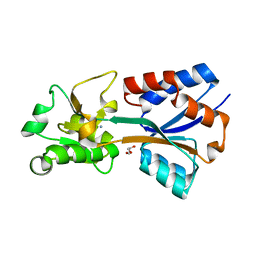 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in ligand-free form | | Descriptor: | AMMONIUM ION, GLYCEROL, Molybdate-binding periplasmic protein, ... | | Authors: | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
7T5A
 
 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in tungstate-bound form | | Descriptor: | AMMONIUM ION, Molybdate-binding periplasmic protein ModA, TUNGSTATE(VI)ION | | Authors: | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | Deposit date: | 2021-12-11 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
7T50
 
 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in chromate-bound form | | Descriptor: | AMMONIUM ION, Chromate, GLYCEROL, ... | | Authors: | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | Deposit date: | 2021-12-11 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
7T51
 
 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in molybdate-bound form | | Descriptor: | AMMONIUM ION, GLYCEROL, MOLYBDATE ION, ... | | Authors: | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | Deposit date: | 2021-12-11 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
