2NX8
 
 | |
3ILB
 
 | |
3IHD
 
 | |
3IHF
 
 | |
3IIH
 
 | |
3IHE
 
 | |
3ILC
 
 | |
1B8J
 
 | | ALKALINE PHOSPHATASE COMPLEXED WITH VANADATE | | Descriptor: | MAGNESIUM ION, PROTEIN (ALKALINE PHOSPHATASE), SULFATE ION, ... | | Authors: | Holtz, K.M, Stec, B, Kantrowitz, E.R. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A model of the transition state in the alkaline phosphatase reaction.
J.Biol.Chem., 274, 1999
|
|
1ALI
 
 | | ALKALINE PHOSPHATASE MUTANT (H412N) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ma, L, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-06-02 | | Release date: | 1995-11-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Escherichia coli alkaline phosphatase: X-ray structural studies of a mutant enzyme (His-412-->Asn) at one of the catalytically important zinc binding sites.
Protein Sci., 4, 1995
|
|
1ALH
 
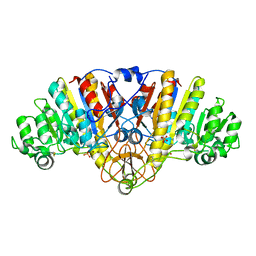 | | KINETICS AND CRYSTAL STRUCTURE OF A MUTANT E. COLI ALKALINE PHOSPHATASE (ASP-369-->ASN): A MECHANISM INVOLVING ONE ZINC PER ACTIVE SITE | | Descriptor: | ALKALINE PHOSPHATASE, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Tibbitts, T.T, Xu, X, Kantrowitz, E.R. | | Deposit date: | 1994-08-23 | | Release date: | 1995-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetics and crystal structure of a mutant Escherichia coli alkaline phosphatase (Asp-369-->Asn): a mechanism involving one zinc per active site.
Protein Sci., 3, 1994
|
|
1ALJ
 
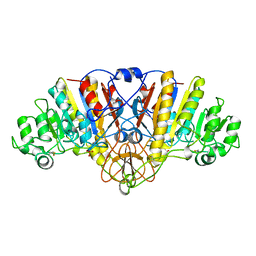 | | ALKALINE PHOSPHATASE MUTANT (H412N) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ma, L, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-06-02 | | Release date: | 1995-11-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Escherichia coli alkaline phosphatase: X-ray structural studies of a mutant enzyme (His-412-->Asn) at one of the catalytically important zinc binding sites.
Protein Sci., 4, 1995
|
|
1ANJ
 
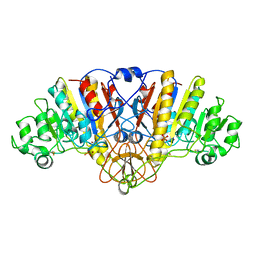 | | ALKALINE PHOSPHATASE (K328H) | | Descriptor: | ALKALINE PHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Murphy, J.E, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-09-06 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations at positions 153 and 328 in Escherichia coli alkaline phosphatase provide insight towards the structure and function of mammalian and yeast alkaline phosphatases.
J.Mol.Biol., 253, 1995
|
|
1ANI
 
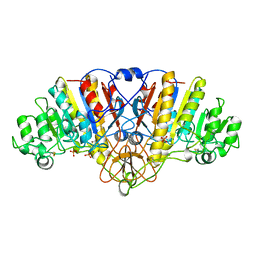 | | ALKALINE PHOSPHATASE (D153H, K328H) | | Descriptor: | ALKALINE PHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Murphy, J.E, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-09-06 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutations at positions 153 and 328 in Escherichia coli alkaline phosphatase provide insight towards the structure and function of mammalian and yeast alkaline phosphatases.
J.Mol.Biol., 253, 1995
|
|
3CMD
 
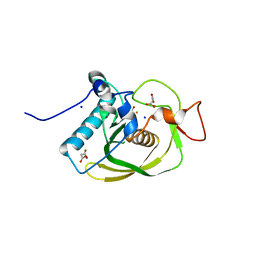 | | Crystal structure of peptide deformylase from VRE-E.faecium | | Descriptor: | FE (III) ION, MALONATE ION, Peptide deformylase, ... | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2008-03-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insight into the antibacterial drug design and architectural mechanism of peptide recognition from the E. faecium peptide deformylase structure.
Proteins, 74, 2009
|
|
3E6E
 
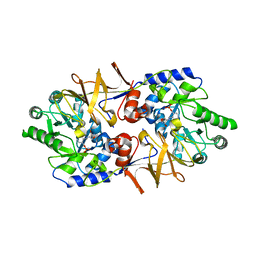 | | Crystal structure of Alanine racemase from E.faecalis complex with cycloserine | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Hwang, K.Y, Priyadarshi, A, Lee, E.H, Sung, M.W. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the alanine racemase from Enterococcus faecalis.
Biochim.Biophys.Acta, 1794, 2009
|
|
3E5P
 
 | | Crystal structure of alanine racemase from E.faecalis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alanine racemase, NONAETHYLENE GLYCOL, ... | | Authors: | Hwang, K.Y, Priyadarshi, A, Lee, E.H, Sung, M.W. | | Deposit date: | 2008-08-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the alanine racemase from Enterococcus faecalis.
Biochim.Biophys.Acta, 1794, 2009
|
|
3GNT
 
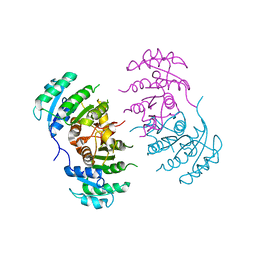 | |
3GNS
 
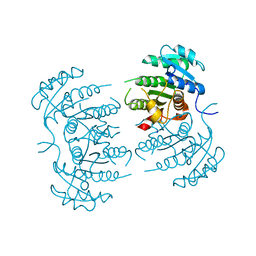 | |
3GR6
 
 | |
3HWX
 
 | |
3HWW
 
 | |
1FKL
 
 | | ATOMIC STRUCTURE OF FKBP12-RAPAYMYCIN, AN IMMUNOPHILIN-IMMUNOSUPPRESSANT COMPLEX | | Descriptor: | FK506 BINDING PROTEIN, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1FKK
 
 | | ATOMIC STRUCTURE OF FKBP12, AN IMMUNOPHILIN BINDING PROTEIN | | Descriptor: | FK506 BINDING PROTEIN, SULFATE ION | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1FKJ
 
 | | ATOMIC STRUCTURE OF FKBP12-FK506, AN IMMUNOPHILIN IMMUNOSUPPRESSANT COMPLEX | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506 BINDING PROTEIN | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
2P6B
 
 | | Crystal Structure of Human Calcineurin in Complex with PVIVIT Peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B isoform 1, Calmodulin-dependent calcineurin A subunit alpha isoform, ... | | Authors: | Li, H, Zhang, L, Rao, A, Harrison, S.C, Hogan, P.G. | | Deposit date: | 2007-03-16 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of calcineurin in complex with PVIVIT peptide: Portrait of a low-affinity signalling interaction
J.Mol.Biol., 369, 2007
|
|
