1J3Q
 
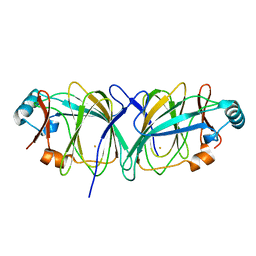 | | Crystal structure of Thermococcus litoralis phosphogrucose isomerase soaked with FeSO4 | | Descriptor: | FE (III) ION, Phosphoglucose Isomerase | | Authors: | Jeong, J.-J, Fushinobu, S, Ito, S, Hidaka, M, Shoun, H, Wakagi, T. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a novel cupin-type phosphoglucose isomerase
To be Published
|
|
4Y61
 
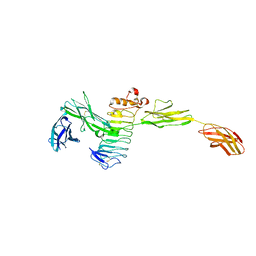 | | Crystal structure of the complex between Slitrk2 LRR1 and PTP delta Ig1-Fn1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase delta, SLIT and NTRK-like protein 2 | | Authors: | Yamgata, A, Sato, Y, Goto-Ito, S, Uemura, T, Maeda, A, Shiroshima, T, Yoshida, T, Fukai, S. | | Deposit date: | 2015-02-12 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.358 Å) | | Cite: | Structure of Slitrk2-PTP delta complex reveals mechanisms for splicing-dependent trans-synaptic adhesion.
Sci Rep, 5, 2015
|
|
5GVC
 
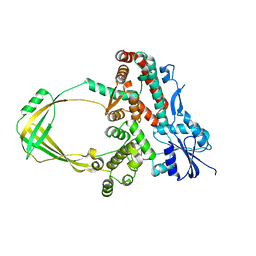 | | Human Topoisomerase IIIb topo domain | | Descriptor: | DNA topoisomerase 3-beta-1, MAGNESIUM ION | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
5H1A
 
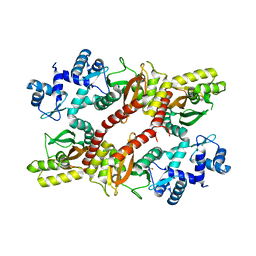 | | Crystal structure of an IclR homolog from Microbacterium sp. strain HM58-2 | | Descriptor: | IclR transcription factor homolog, PHOSPHATE ION | | Authors: | Akiyama, T, Yamada, Y, Takaya, N, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an IclR homologue from Microbacterium sp. strain HM58-2.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5GVD
 
 | | Human TDRD3 DUF1767-OB domains | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
5GVE
 
 | | Human TOP3B-TDRD3 complex | | Descriptor: | DNA topoisomerase 3-beta-1, MAGNESIUM ION, Tudor domain-containing protein 3 | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.606 Å) | | Cite: | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
1WMF
 
 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (oxidized form, 1.73 angstrom) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1WME
 
 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.50 angstrom, 293 K) | | Descriptor: | CALCIUM ION, protease | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1WMD
 
 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.30 angstrom, 100 K) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
5ZQU
 
 | | Crystal structure of tetrameric RXRalpha-LBD complexed with partial agonist CBt-PMN | | Descriptor: | 1-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)benzotriazole-5-carboxylic acid, BROMIDE ION, Retinoic acid receptor RXR-alpha | | Authors: | Miyashita, Y, Numoto, N, Arulmozhiraja, S, Nakano, S, Matsuo, N, Shimizu, K, Kakuta, H, Ito, S, Ikura, T, Ito, N, Tokiwa, H. | | Deposit date: | 2018-04-20 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.60038781 Å) | | Cite: | Dual conformation of the ligand induces the partial agonistic activity of retinoid X receptor alpha (RXR alpha ).
FEBS Lett., 593, 2019
|
|
6L96
 
 | | Structure of PPARalpha-LBD/pemafibrate/SRC1 peptide | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor alpha, SRC1 coactivator peptide | | Authors: | Kawasaki, M, Kambe, A, Yamamoto, Y, Arulmozhira, S, Ito, S, Nakagawa, Y, Tokiwa, H, Nakano, S, Shimano, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Elucidation of Molecular Mechanism of a Selective PPAR alpha Modulator, Pemafibrate, through Combinational Approaches of X-ray Crystallography, Thermodynamic Analysis, and First-Principle Calculations.
Int J Mol Sci, 21, 2020
|
|
6M5W
 
 | | Co-crystal structure of human serine hydroxymethyltransferase 1 in complex with Pyridoxal 5'-phosphate (PLP) and glycodeoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Ota, T, Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K, Sando, S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for selective inhibition of human serine hydroxymethyltransferase by secondary bile acid conjugate.
Iscience, 24, 2021
|
|
6M5O
 
 | | Co-crystal structure of human serine hydroxymethyltransferase 2 in complex with Pyridoxal 5'-phosphate (PLP) and glycodeoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCINE, Serine hydroxymethyltransferase, ... | | Authors: | Ota, T, Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K, Sando, S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.30000663 Å) | | Cite: | Structural basis for selective inhibition of human serine hydroxymethyltransferase by secondary bile acid conjugate.
Iscience, 24, 2021
|
|
7C86
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: Dark state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RETINAL, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-05-28 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
6JNO
 
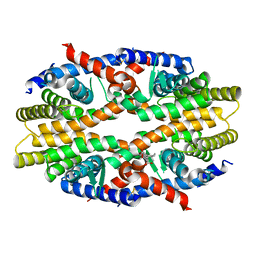 | | RXRa structure complexed with CU-6PMN | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-17 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Competitive Binding Assay with an Umbelliferone-Based Fluorescent Rexinoid for Retinoid X Receptor Ligand Screening.
J.Med.Chem., 62, 2019
|
|
6JNR
 
 | | RXRa structure complexed with CU-6PMN and SRC1 peptide. | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RXRa structure complexed with CU-6PMN and SRC1 peptide.
To Be Published
|
|
6KTK
 
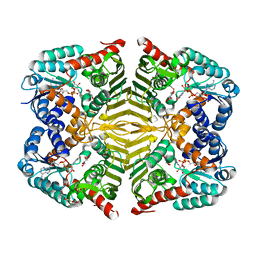 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NADH and L-glucono-1,5-lactone, from Paracoccus laeviglucosivorans | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6KTL
 
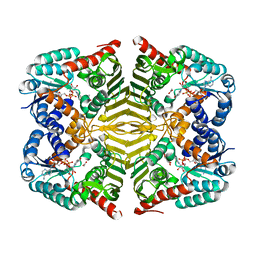 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NAD and myo-inositol, from Paracoccus laeviglucosivorans | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6KTJ
 
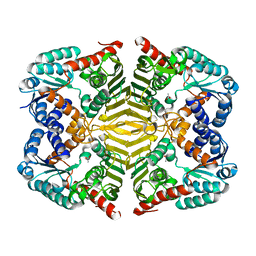 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, apo-form, from Paracoccus laeviglucosivorans | | Descriptor: | ACETATE ION, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6LU2
 
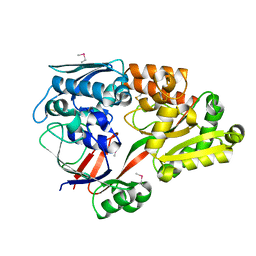 | | Crystal structure of a substrate binding protein from Microbacterium hydrocarbonoxydans | | Descriptor: | Substrate binding protein | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6LU3
 
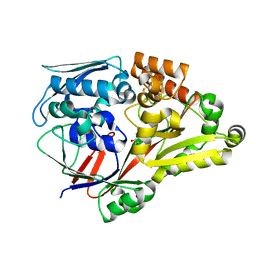 | | Crystal structure of a substrate binding protein from Microbacterium hydrocarbonoxydans complexed with 4-hydroxybenzoate hydrazide | | Descriptor: | 4-oxidanylbenzohydrazide, Substrate binding protein | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6LU4
 
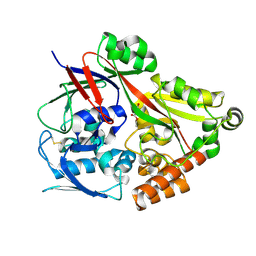 | | Crystal structure of the substrate binding protein from Microbacterium hydrocarbonoxydans complexed with propylparaben | | Descriptor: | Substrate binding protein, propyl 4-hydroxybenzoate | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
7BXS
 
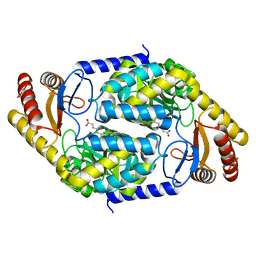 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator glycine binding form | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXQ
 
 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator L-Threonine binding form | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-allothreonine | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXP
 
 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
