3JRR
 
 | | Crystal structure of the ligand binding suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor | | 分子名称: | Inositol 1,4,5-trisphosphate receptor type 3 | | 著者 | Chan, J, Ishiyama, N, Ikura, M. | | 登録日 | 2009-09-08 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A 1.9 angstrom crystal structure of the suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor
To be Published
|
|
3L6Y
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | 分子名称: | Catenin delta-1, E-cadherin | | 著者 | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | 登録日 | 2009-12-27 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3L6X
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | 分子名称: | Catenin delta-1, E-cadherin, SULFATE ION | | 著者 | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | 登録日 | 2009-12-27 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
1SUH
 
 | | AMINO-TERMINAL DOMAIN OF EPITHELIAL CADHERIN IN THE CALCIUM BOUND STATE, NMR, 20 STRUCTURES | | 分子名称: | EPITHELIAL CADHERIN | | 著者 | Overduin, M, Tong, K.I, Kay, C.M, Ikura, M. | | 登録日 | 1996-01-30 | | 公開日 | 1996-07-11 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 1H, 15N and 13C resonance assignments and monomeric structure of the amino-terminal extracellular domain of epithelial cadherin.
J.Biomol.NMR, 7, 1996
|
|
1PRR
 
 | | NMR-DERIVED THREE-DIMENSIONAL SOLUTION STRUCTURE OF PROTEIN S COMPLEXED WITH CALCIUM | | 分子名称: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | 著者 | Bagby, S, Harvey, T.S, Eagle, S.G, Inouye, S, Ikura, M. | | 登録日 | 1994-03-25 | | 公開日 | 1994-08-31 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR-derived three-dimensional solution structure of protein S complexed with calcium.
Structure, 2, 1994
|
|
1PRS
 
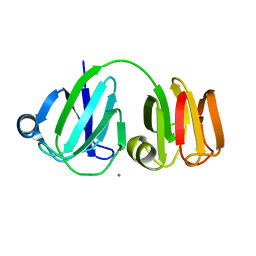 | | NMR-DERIVED THREE-DIMENSIONAL SOLUTION STRUCTURE OF PROTEIN S COMPLEXED WITH CALCIUM | | 分子名称: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | 著者 | Bagby, S, Harvey, T.S, Eagle, S.G, Inouye, S, Ikura, M. | | 登録日 | 1994-03-25 | | 公開日 | 1994-08-31 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR-derived three-dimensional solution structure of protein S complexed with calcium.
Structure, 2, 1994
|
|
4O25
 
 | | Structure of Wild Type Mus musculus Rheb bound to GTP | | 分子名称: | GTP-binding protein Rheb, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | 著者 | Mazhab-Jafari, M.T, Marshall, C.B, Ho, J, Ishiyama, N, Stambolic, V, Ikura, M. | | 登録日 | 2013-12-16 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR).
J.Biol.Chem., 289, 2014
|
|
4O2L
 
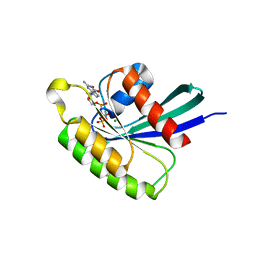 | | Structure of Mus musculus Rheb G63A mutant bound to GTP | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GTP-binding protein Rheb, ... | | 著者 | Mazhab-Jafari, M.T, Marshall, C.B, Ho, J, Ishiyama, N, Stambolic, V, Ikura, M. | | 登録日 | 2013-12-17 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR).
J.Biol.Chem., 289, 2014
|
|
4O2R
 
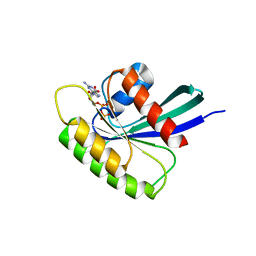 | | Structure of Mus musculus Rheb G63V mutant bound to GDP | | 分子名称: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Mazhab-Jafari, M.T, Marshall, C.B, Ho, J, Ishiyama, N, Stambolic, V, Ikura, M. | | 登録日 | 2013-12-17 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR).
J.Biol.Chem., 289, 2014
|
|
6AMB
 
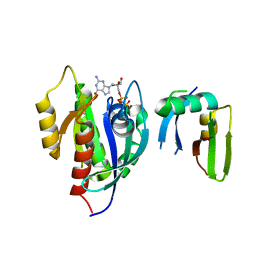 | | Crystal Structure of the Afadin RA1 domain in complex with HRAS | | 分子名称: | Afadin, GTPase HRas, MAGNESIUM ION, ... | | 著者 | Smith, M.J, Ishiyama, N, Ikura, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-11-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Evolution of AF6-RAS association and its implications in mixed-lineage leukemia.
Nat Commun, 8, 2017
|
|
2GW3
 
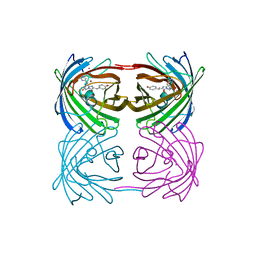 | | Crystal structure of stony coral fluorescent protein Kaede, green form | | 分子名称: | Kaede, NICKEL (II) ION | | 著者 | Hayashi, I, Mizuno, H, Miyawaki, A, Ikura, M. | | 登録日 | 2006-05-03 | | 公開日 | 2007-05-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystallographic evidence for water-assisted photo-induced peptide cleavage in the stony coral fluorescent protein Kaede.
J.Mol.Biol., 372, 2007
|
|
2GW4
 
 | | Crystal structure of stony coral fluorescent protein Kaede, red form | | 分子名称: | Kaede, NICKEL (II) ION | | 著者 | Hayashi, I, Mizuno, H, Miyawako, A, Ikura, M. | | 登録日 | 2006-05-03 | | 公開日 | 2007-05-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystallographic evidence for water-assisted photo-induced peptide cleavage in the stony coral fluorescent protein Kaede.
J.Mol.Biol., 372, 2007
|
|
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | 分子名称: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | 著者 | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | 登録日 | 2018-02-06 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
1JOY
 
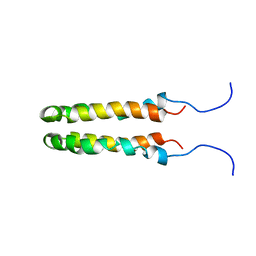 | | SOLUTION STRUCTURE OF THE HOMODIMERIC DOMAIN OF ENVZ FROM ESCHERICHIA COLI BY MULTI-DIMENSIONAL NMR. | | 分子名称: | PROTEIN (ENVZ_ECOLI) | | 著者 | Tomomori, C, Tanaka, T, Dutta, R, Park, H, Saha, S.K, Zhu, Y, Ishima, R, Liu, D, Tong, K.I, Kurokawa, H, Qian, H, Inouye, M, Ikura, M. | | 登録日 | 1998-12-28 | | 公開日 | 2000-01-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ.
Nat.Struct.Biol., 6, 1999
|
|
6CCX
 
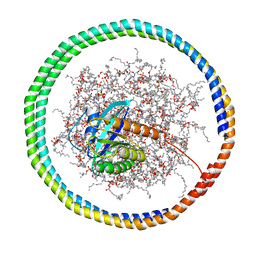 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | 分子名称: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | 著者 | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | 登録日 | 2018-02-07 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | 登録日 | 2018-02-07 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6DUW
 
 | |
6DV1
 
 | |
6DUY
 
 | |
1N4K
 
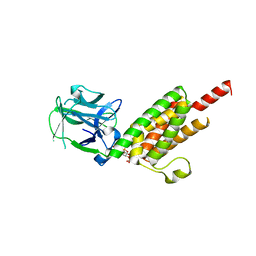 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with IP3 | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | 著者 | Bosanac, I, Alattia, J.R, Mal, T.K, Chan, J, Talarico, S, Tong, F.K, Tong, K.I, Yoshikawa, F, Furuichi, T, Iwai, M, Michikawa, T, Mikoshiba, K, Ikura, M. | | 登録日 | 2002-10-31 | | 公開日 | 2002-12-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the inositol 1,4,5-trisphosphate receptor
binding core in complex with its ligand.
Nature, 420, 2002
|
|
1MUX
 
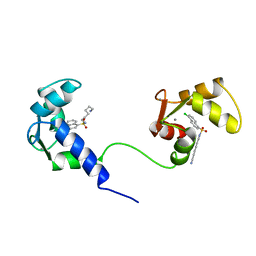 | | SOLUTION NMR STRUCTURE OF CALMODULIN/W-7 COMPLEX: THE BASIS OF DIVERSITY IN MOLECULAR RECOGNITION, 30 STRUCTURES | | 分子名称: | CALCIUM ION, CALMODULIN, N-(6-AMINOHEXYL)-5-CHLORO-1-NAPHTHALENESULFONAMIDE | | 著者 | Osawa, M, Swindells, M.B, Tanikawa, J, Tanaka, T, Mase, T, Furuya, T, Ikura, M. | | 登録日 | 1997-09-06 | | 公開日 | 1998-10-14 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of calmodulin-W-7 complex: the basis of diversity in molecular recognition.
J.Mol.Biol., 276, 1998
|
|
1TFB
 
 | |
1IKU
 
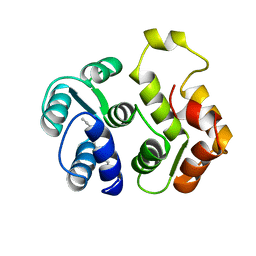 | | myristoylated recoverin in the calcium-free state, NMR, 22 structures | | 分子名称: | MYRISTIC ACID, RECOVERIN | | 著者 | Tanaka, T, Ames, J.B, Harvey, T.S, Stryer, L, Ikura, M. | | 登録日 | 1996-01-18 | | 公開日 | 1996-07-11 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state.
Nature, 376, 1995
|
|
1XZZ
 
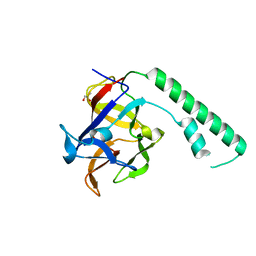 | | Crystal structure of the ligand binding suppressor domain of type 1 inositol 1,4,5-trisphosphate receptor | | 分子名称: | GLYCEROL, Inositol 1,4,5-trisphosphate receptor type 1 | | 著者 | Bosanac, I, Yamazaki, H, Matsu-ura, T, Michikawa, T, Mikoshiba, K, Ikura, M. | | 登録日 | 2004-11-13 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the ligand binding suppressor domain of type 1 inositol 1,4,5-trisphosphate receptor.
Mol.Cell, 17, 2005
|
|
1RO4
 
 | |
