2MSE
 
 | | NMR data-driven model of GTPase KRas-GNP:ARafRBD complex tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSC
 
 | | NMR data-driven model of GTPase KRas-GDP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-09-05 | | Last modified: | 2019-03-27 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
2K29
 
 | |
2KC8
 
 | | Structure of E. coli toxin RelE (R81A/R83A) mutant in complex with antitoxin RelBc (K47-L79) peptide | | Descriptor: | Antitoxin RelB, Toxin relE | | Authors: | Li, G, Zhang, Y, Inouye, M, Ikura, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-17 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site.
J.Biol.Chem., 284, 2009
|
|
2KC9
 
 | |
2KUO
 
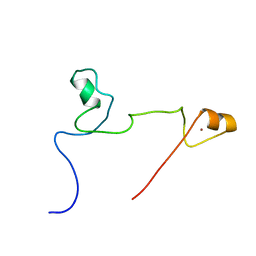 | | Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Li, G.Y, McCulloch, R.D, Fenton, A, Cheung, M, Meng, L, Ikura, M, Koch, C.A. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-05 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and identification of ADP-ribose recognition motifs of aprataxin PNK-like factor (APLF) required for the interaction with sites of DNA damage response
To be Published
|
|
2LQH
 
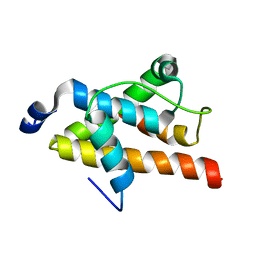 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LQI
 
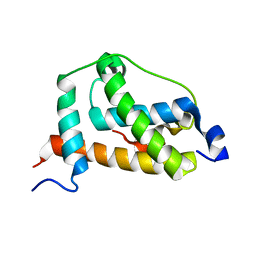 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2l3b conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2MC2
 
 | |
2GW3
 
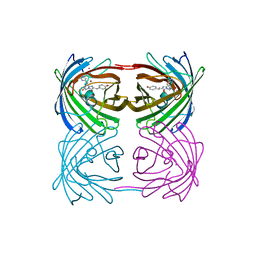 | | Crystal structure of stony coral fluorescent protein Kaede, green form | | Descriptor: | Kaede, NICKEL (II) ION | | Authors: | Hayashi, I, Mizuno, H, Miyawaki, A, Ikura, M. | | Deposit date: | 2006-05-03 | | Release date: | 2007-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic evidence for water-assisted photo-induced peptide cleavage in the stony coral fluorescent protein Kaede.
J.Mol.Biol., 372, 2007
|
|
2GW4
 
 | | Crystal structure of stony coral fluorescent protein Kaede, red form | | Descriptor: | Kaede, NICKEL (II) ION | | Authors: | Hayashi, I, Mizuno, H, Miyawako, A, Ikura, M. | | Deposit date: | 2006-05-03 | | Release date: | 2007-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic evidence for water-assisted photo-induced peptide cleavage in the stony coral fluorescent protein Kaede.
J.Mol.Biol., 372, 2007
|
|
1MYW
 
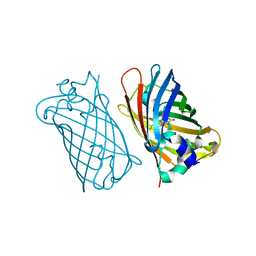 | | CRYSTAL STRUCTURE OF A YELLOW FLUORESCENT PROTEIN WITH IMPROVED MATURATION AND REDUCED ENVIRONMENTAL SENSITIVITY | | Descriptor: | Green fluorescent protein | | Authors: | Rekas, A, Alattia, J.R, Nagai, T, Miyawaki, A, Ikura, M. | | Deposit date: | 2002-10-04 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Venus, a Yellow Fluorescent
Protein with Improved Maturation and
Reduced Environmental Sensitivity
J.Biol.Chem., 277, 2002
|
|
1NWD
 
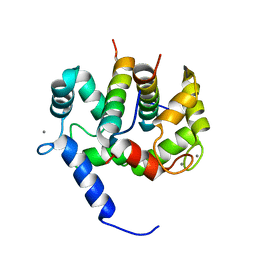 | | Solution Structure of Ca2+/Calmodulin bound to the C-terminal Domain of Petunia Glutamate Decarboxylase | | Descriptor: | CALCIUM ION, Calmodulin, Glutamate decarboxylase | | Authors: | Yap, K.L, Yuan, T, Mal, T.K, Vogel, H.J, Ikura, M. | | Deposit date: | 2003-02-06 | | Release date: | 2003-04-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Simultaneous Binding of Two Carboxy-terminal Peptides
of Plant Glutamate Decarboxylase to Calmodulin
J.Mol.Biol., 328, 2003
|
|
1PA7
 
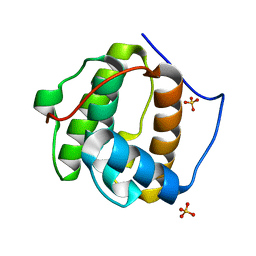 | |
1TBA
 
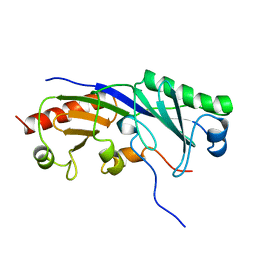 | | SOLUTION STRUCTURE OF A TBP-TAFII230 COMPLEX: PROTEIN MIMICRY OF THE MINOR GROOVE SURFACE OF THE TATA BOX UNWOUND BY TBP, NMR, 25 STRUCTURES | | Descriptor: | TRANSCRIPTION INITIATION FACTOR IID 230K CHAIN, TRANSCRIPTION INITIATION FACTOR TFIID | | Authors: | Liu, D, Ishima, R, Tong, K.I, Bagby, S, Kokubo, T, Muhandiram, D.R, Kay, L.E, Nakatani, Y, Ikura, M. | | Deposit date: | 1998-08-16 | | Release date: | 1999-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP.
Cell(Cambridge,Mass.), 94, 1998
|
|
1TXQ
 
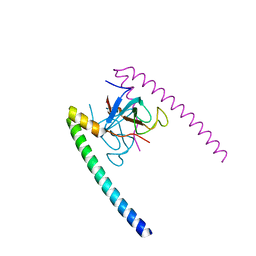 | |
1UEG
 
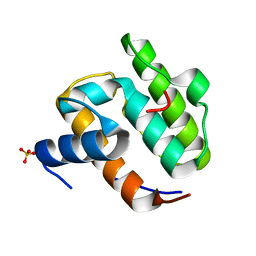 | |
1XZZ
 
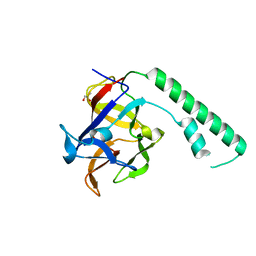 | | Crystal structure of the ligand binding suppressor domain of type 1 inositol 1,4,5-trisphosphate receptor | | Descriptor: | GLYCEROL, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Bosanac, I, Yamazaki, H, Matsu-ura, T, Michikawa, T, Mikoshiba, K, Ikura, M. | | Deposit date: | 2004-11-13 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ligand binding suppressor domain of type 1 inositol 1,4,5-trisphosphate receptor.
Mol.Cell, 17, 2005
|
|
2H7B
 
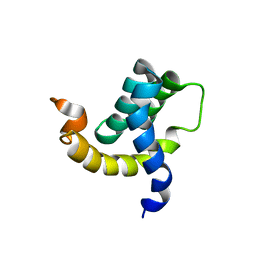 | | Solution structure of the eTAFH domain from the human leukemia-associated fusion protein AML1-ETO | | Descriptor: | Core-binding factor, ML1-ETO | | Authors: | Plevin, M.J, Zhang, J, Guo, C, Roeder, R.G, Ikura, M. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-11 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The acute myeloid leukemia fusion protein AML1-ETO targets E proteins via a paired amphipathic helix-like TBP-associated factor homology domain
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2HQH
 
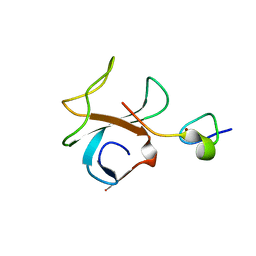 | | Crystal structure of p150Glued and CLIP-170 | | Descriptor: | Dynactin-1, Restin, ZINC ION | | Authors: | Hayashi, I, Ikura, M. | | Deposit date: | 2006-07-18 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CLIP170 autoinhibition mimics intermolecular interactions with p150Glued or EB1.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2IB5
 
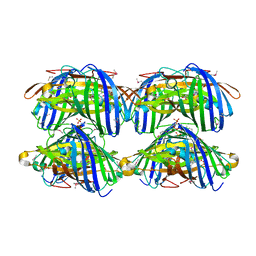 | |
2IB6
 
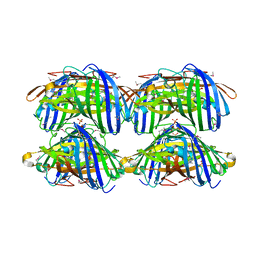 | | Structural characterization of a blue chromoprotein and its yellow mutant from the sea anemone cnidopus japonicus | | Descriptor: | PHOSPHATE ION, Yellow mutant chromo protein | | Authors: | Chan, M.C.Y, Bosanac, I, Ho, D, Prive, G, Ikura, M. | | Deposit date: | 2006-09-10 | | Release date: | 2006-10-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of a Blue Chromoprotein and Its Yellow Mutant from the Sea Anemone Cnidopus Japonicus
J.Biol.Chem., 281, 2006
|
|
2K86
 
 | | Solution Structure of FOXO3a Forkhead domain | | Descriptor: | Forkhead box protein O3 | | Authors: | Wang, F, Marshall, C.B, Li, G, Plevin, M.J, Ikura, M. | | Deposit date: | 2008-09-02 | | Release date: | 2008-10-14 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural characterization of an intramolecular interaction in FOXO3a and its binding with p53.
J.Mol.Biol., 384, 2008
|
|
