1YOM
 
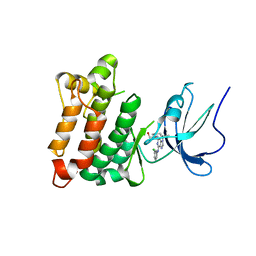 | | Crystal structure of Src kinase domain in complex with Purvalanol A | | Descriptor: | 2-({6-[(3-CHLOROPHENYL)AMINO]-9-ISOPROPYL-9H-PURIN-2-YL}AMINO)-3-METHYLBUTAN-1-OL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Breitenlechner, C.B, Kairies, N.A, Honold, K, Scheiblich, S, Koll, H, Greiter, E, Koch, S, Schaefer, W, Huber, R, Engh, R.A. | | Deposit date: | 2005-01-27 | | Release date: | 2006-01-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of active SRC kinase domain complexes
J.Mol.Biol., 353, 2005
|
|
1ZLI
 
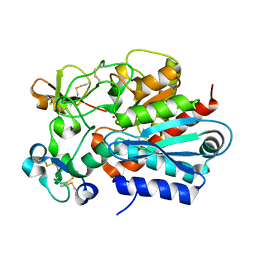 | | Crystal structure of the tick carboxypeptidase inhibitor in complex with human carboxypeptidase B | | Descriptor: | Carboxypeptidase B, ZINC ION, carboxypeptidase inhibitor | | Authors: | Arolas, J.L, Popowicz, G.M, Lorenzo, J, Sommerhoff, C.P, Huber, R, Aviles, F.X, Holak, T.A. | | Deposit date: | 2005-05-06 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Three-Dimensional Structures of Tick Carboxypeptidase Inhibitor in Complex with A/B Carboxypeptidases Reveal a Novel Double-headed Binding Mode
J.Mol.Biol., 350, 2005
|
|
1ZLH
 
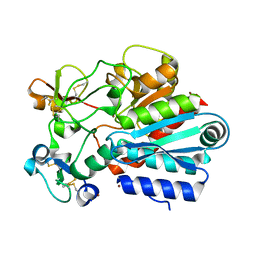 | | Crystal structure of the tick carboxypeptidase inhibitor in complex with bovine carboxypeptidase A | | Descriptor: | Carboxypeptidase A1, ZINC ION, carboxypeptidase inhibitor | | Authors: | Arolas, J.L, Popowicz, G.M, Lorenzo, J, Sommerhoff, C.P, Huber, R, Aviles, F.X, Holak, T.A. | | Deposit date: | 2005-05-06 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Three-Dimensional Structures of Tick Carboxypeptidase Inhibitor in Complex with A/B Carboxypeptidases Reveal a Novel Double-headed Binding Mode
J.Mol.Biol., 350, 2005
|
|
2ACH
 
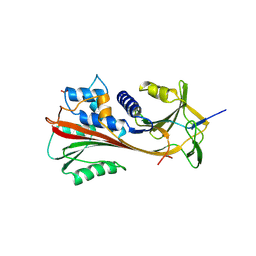 | | CRYSTAL STRUCTURE OF CLEAVED HUMAN ALPHA1-ANTICHYMOTRYPSIN AT 2.7 ANGSTROMS RESOLUTION AND ITS COMPARISON WITH OTHER SERPINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA 1-ANTICHYMOTRYPSIN, PHOSPHATE ION, ... | | Authors: | Baumann, U, Huber, R, Bode, W, Grosse, D, Lesjak, M, Laurell, C.B. | | Deposit date: | 1993-04-26 | | Release date: | 1993-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cleaved human alpha 1-antichymotrypsin at 2.7 A resolution and its comparison with other serpins.
J.Mol.Biol., 218, 1991
|
|
1ONP
 
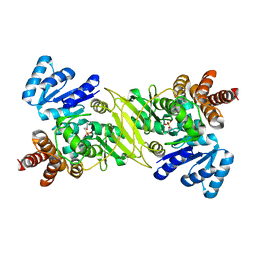 | | IspC complex with Mn2+ and fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION | | Authors: | Steinbacher, S, Kaiser, J, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol
4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria
drug development.
J.BIOL.CHEM., 278, 2003
|
|
1P3Y
 
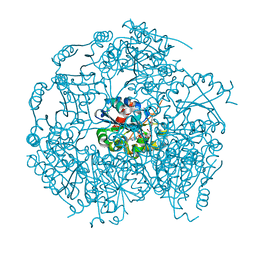 | | MrsD from Bacillus sp. HIL-Y85/54728 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MrsD protein | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbacher, S. | | Deposit date: | 2003-04-19 | | Release date: | 2003-08-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of MrsD, an FAD-binding protein of the HFCD family.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1JD2
 
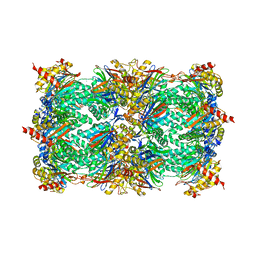 | | Crystal Structure of the yeast 20S Proteasome:TMC-95A complex: A non-covalent Proteasome Inhibitor | | Descriptor: | MAGNESIUM ION, PROTEASOME COMPONENT C1, PROTEASOME COMPONENT C11, ... | | Authors: | Groll, M, Koguchi, Y, Huber, R, Kohno, J. | | Deposit date: | 2001-06-12 | | Release date: | 2002-02-13 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the 20 S proteasome:TMC-95A complex: a non-covalent proteasome inhibitor.
J.Mol.Biol., 311, 2001
|
|
1P0N
 
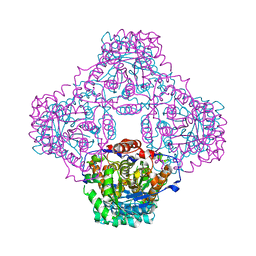 | | IPP:DMAPP isomerase type II, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Isopentenyl-diphosphate delta-isomerase | | Authors: | Steinbacher, S, Kaiser, J, Gerhardt, S, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Type II Isopentenyl Diphosphate:Dimethylallyl Diphosphate Isomerase from Bacillus subtilis
J.Mol.Biol., 329, 2003
|
|
1JQG
 
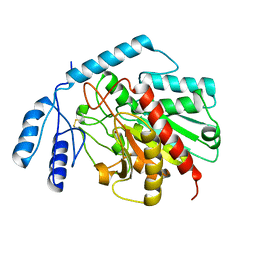 | | Crystal Structure of the Carboxypeptidase A from Helicoverpa Armigera | | Descriptor: | ZINC ION, carboxypeptidase A | | Authors: | Estebanez-Perpina, E, Bayes, A, Vendrell, J, Jongsma, M.A, Bown, D.P, Gatehouse, J.A, Huber, R, Bode, W, Aviles, F.X, Reverter, D. | | Deposit date: | 2001-08-07 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a novel mid-gut procarboxypeptidase from the cotton pest Helicoverpa armigera.
J.Mol.Biol., 313, 2001
|
|
1JED
 
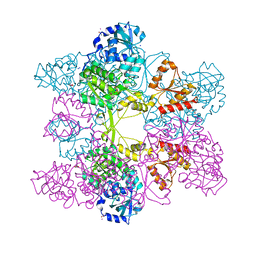 | | Crystal Structure of ATP Sulfurylase in complex with ADP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ullrich, T.C, Huber, R. | | Deposit date: | 2001-06-17 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The complex structures of ATP sulfurylase with thiosulfate, ADP and chlorate reveal new insights in inhibitory effects and the catalytic cycle.
J.Mol.Biol., 313, 2001
|
|
1JG5
 
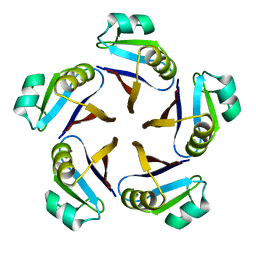 | | CRYSTAL STRUCTURE OF RAT GTP CYCLOHYDROLASE I FEEDBACK REGULATORY PROTEIN, GFRP | | Descriptor: | GTP CYCLOHYDROLASE I FEEDBACK REGULATORY PROTEIN, POTASSIUM ION | | Authors: | Bader, G, Schiffmann, S, Herrmann, A, Fischer, M, Gutlich, M, Auerbach, G, Ploom, T, Bacher, A, Huber, R, Lemm, T. | | Deposit date: | 2001-06-23 | | Release date: | 2001-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of rat GTP cyclohydrolase I feedback regulatory protein, GFRP.
J.Mol.Biol., 312, 2001
|
|
1JHJ
 
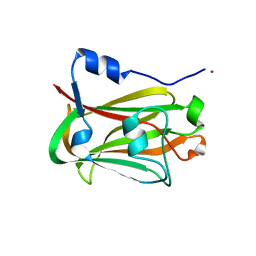 | | Crystal structure of the APC10/Doc1 subunit of the human anaphase-promoting complex | | Descriptor: | APC10, NICKEL (II) ION | | Authors: | Wendt, K.S, Vodermaier, H.C, Jacob, U, Gieffers, C, Gmachl, M, Peters, J.-M, Huber, R, Sondermann, P. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-24 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex
Nat.Struct.Biol., 8, 2001
|
|
1JEC
 
 | |
1PIX
 
 | | Crystal structure of the carboxyltransferase subunit of the bacterial ion pump glutaconyl-coenzyme A decarboxylase | | Descriptor: | FORMIC ACID, Glutaconyl-CoA decarboxylase A subunit, SULFATE ION | | Authors: | Wendt, K.S, Schall, I, Huber, R, Buckel, W, Jacob, U. | | Deposit date: | 2003-05-30 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the carboxyltransferase subunit of the bacterial sodium ion pump glutaconyl-coenzyme A decarboxylase
Embo J., 22, 2003
|
|
1JEE
 
 | | Crystal Structure of ATP Sulfurylase in complex with chlorate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Ullrich, T.C, Huber, R. | | Deposit date: | 2001-06-17 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The complex structures of ATP sulfurylase with thiosulfate, ADP and chlorate reveal new insights in inhibitory effects and the catalytic cycle.
J.Mol.Biol., 313, 2001
|
|
1PKQ
 
 | | Myelin Oligodendrocyte Glycoprotein-(8-18C5) Fab-complex | | Descriptor: | (8-18C5) chimeric Fab, heavy chain, light chain, ... | | Authors: | Breithaupt, C, Schubart, A, Zander, H, Skerra, A, Huber, R, Linington, C, Jacob, U. | | Deposit date: | 2003-06-06 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the antigenicity of myelin oligodendrocyte glycoprotein
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1K32
 
 | |
1PKO
 
 | | Myelin Oligodendrocyte Glycoprotein (MOG) | | Descriptor: | Myelin Oligodendrocyte Glycoprotein | | Authors: | Breithaupt, C, Schubart, A, Zander, H, Skerra, A, Huber, R, Linington, C, Jacob, U. | | Deposit date: | 2003-06-06 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the antigenicity of myelin oligodendrocyte glycoprotein
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1KLI
 
 | | Cofactor-and substrate-assisted activation of factor VIIa | | Descriptor: | BENZAMIDINE, CALCIUM ION, GLYCEROL, ... | | Authors: | Sichler, K, Banner, D.W, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structure of Uninhibited Factor VIIa Link its Cofactor and Substrate-assisted Activation to Specific Interactions
J.Mol.Biol., 322, 2002
|
|
1KFX
 
 | | Crystal Structure of Human m-Calpain Form I | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1KL7
 
 | | Crystal Structure of Threonine Synthase from Yeast | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Threonine Synthase | | Authors: | Garrido-Franco, M, Ehlert, S, Messerschmidt, A, Marinkovic, S, Huber, R, Laber, B, Bourenkov, G.P, Clausen, T. | | Deposit date: | 2001-12-11 | | Release date: | 2002-04-24 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of threonine synthase from yeast.
J.Biol.Chem., 277, 2002
|
|
1KFU
 
 | | Crystal Structure of Human m-Calpain Form II | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1KLJ
 
 | | Crystal structure of uninhibited factor VIIa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, factor VIIa | | Authors: | Sichler, K, Banner, D, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of uninhibited factor VIIa link its cofactor and substrate-assisted activation to specific interactions.
J.Mol.Biol., 322, 2002
|
|
1QDB
 
 | | CYTOCHROME C NITRITE REDUCTASE | | Descriptor: | CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, HEME C, ... | | Authors: | Einsle, O, Messerschmidt, A, Stach, P, Huber, R, Kroneck, P.M.H. | | Deposit date: | 1999-05-19 | | Release date: | 1999-08-18 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of cytochrome c nitrite reductase.
Nature, 400, 1999
|
|
1KZ6
 
 | | Mutant enzyme W63Y/L119F Lumazine Synthase from S.pombe | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
