1CJ3
 
 | | MUTANT TYR38GLU OF PARA-HYDROXYBENZOATE HYDROXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Overkamp, K.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-04-21 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase.
J.Mol.Biol., 292, 1999
|
|
1CJ4
 
 | | MUTANT Q34T OF PARA-HYDROXYBENZOATE HYDROXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Overkamp, K.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-04-21 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase.
J.Mol.Biol., 292, 1999
|
|
2I6V
 
 | | PDZ domain of EpsC from Vibrio cholerae, residues 219-305 | | Descriptor: | General secretion pathway protein C | | Authors: | Korotkov, K.V, Krumm, B, Bagdasarian, M, Hol, W.G.J. | | Deposit date: | 2006-08-29 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and Functional Studies of EpsC, a Crucial Component of the Type 2 Secretion System from Vibrio cholerae.
J.Mol.Biol., 363, 2006
|
|
2I4S
 
 | | PDZ domain of EpsC from Vibrio cholerae, residues 204-305 | | Descriptor: | General secretion pathway protein C | | Authors: | Korotkov, K.V, Krumm, B, Bagdasarian, M, Hol, W.G.J. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-17 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and Functional Studies of EpsC, a Crucial Component of the Type 2 Secretion System from Vibrio cholerae.
J.Mol.Biol., 363, 2006
|
|
2ORA
 
 | | RHODANESE (THIOSULFATE: CYANIDE SULFURTRANSFERASE) | | Descriptor: | OXIDIZED RHODANESE | | Authors: | Gliubich, F, Gazerro, M, Zanotti, G, Delbono, S, Berni, R. | | Deposit date: | 1996-02-22 | | Release date: | 1996-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Active site structural features for chemically modified forms of rhodanese.
J.Biol.Chem., 271, 1996
|
|
2X0N
 
 | | Structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei determined from Laue data | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, GLYCOSOMAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Vellieux, F.M.D, Hajdu, J, Hol, W.G.J. | | Deposit date: | 2009-12-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma Brucei Determined from Laue Data.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
6VMT
 
 | |
3C1Q
 
 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Abendroth, J, Mitchell, D.D, Korotkov, K.V, Kreeger, A, Hol, W.G.J. | | Deposit date: | 2008-01-24 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae
J.Struct.Biol., 166, 2009
|
|
3C57
 
 | |
3C3W
 
 | | Crystal Structure of the Mycobacterium tuberculosis Hypoxic Response Regulator DosR | | Descriptor: | SULFATE ION, TWO COMPONENT TRANSCRIPTIONAL REGULATORY PROTEIN DEVR | | Authors: | Wisedchaisri, G, Wu, M, Sherman, D.R, Hol, W.G.J. | | Deposit date: | 2008-01-28 | | Release date: | 2008-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the response regulator DosR from Mycobacterium tuberculosis suggest a helix rearrangement mechanism for phosphorylation activation
J.Mol.Biol., 378, 2008
|
|
3CVL
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphofructokinase (PFK) PTS1 peptide | | Descriptor: | Peroxisome targeting signal 1 receptor PEX5, T. brucei PFK PTS1 peptide Ac-HEELAKL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CVQ
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (7-SKL) | | Descriptor: | GLYCEROL, PTS1 peptide 7-SKL (Ac-SNRWSKL), Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
2PHH
 
 | |
4M84
 
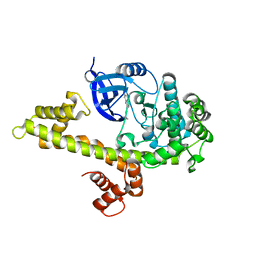 | |
2CHB
 
 | | CHOLERA TOXIN B-PENTAMER COMPLEXED WITH GM1 PENTASACCHARIDE | | Descriptor: | CHOLERA TOXIN, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 1997-06-03 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of receptor binding by cholera toxin mutants.
Protein Sci., 6, 1997
|
|
1DTW
 
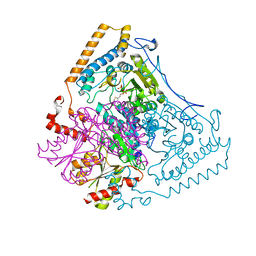 | | HUMAN BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE | | Descriptor: | BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE ALPHA SUBUNIT, BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE BETA SUBUNIT, MAGNESIUM ION, ... | | Authors: | AEvarsson, A, Chuang, J.L, Wynn, R.M, Turley, S, Chuang, D.T, Hol, W.G.J. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease.
Structure Fold.Des., 8, 2000
|
|
1EPX
 
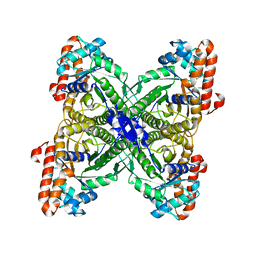 | | CRYSTAL STRUCTURE ANALYSIS OF ALDOLASE FROM L. MEXICANA | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE | | Authors: | Chudzik, D.M, Michels, P.A, de Walque, S, Hol, W.G.J. | | Deposit date: | 2000-03-29 | | Release date: | 2000-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of type 2 peroxisomal targeting signals in two trypanosomatid aldolases.
J.Mol.Biol., 300, 2000
|
|
1EBD
 
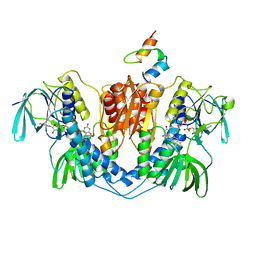 | | DIHYDROLIPOAMIDE DEHYDROGENASE COMPLEXED WITH THE BINDING DOMAIN OF THE DIHYDROLIPOAMIDE ACETYLASE | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE, DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Mande, S.S, Sarfaty, S, Allen, M.D, Perham, R.N, Hol, W.G.J. | | Deposit date: | 1996-02-03 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase.
Structure, 4, 1996
|
|
1EYE
 
 | | 1.7 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF 6-HYDROXYMETHYL-7,8-DIHYDROPTEROATE SYNTHASE (DHPS) FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH 6-HYDROXYMETHYLPTERIN MONOPHOSPHATE | | Descriptor: | DIHYDROPTEROATE SYNTHASE I, MAGNESIUM ION, PTERIN-6-YL-METHYL-MONOPHOSPHATE | | Authors: | Baca, A.M, Sirawaraporn, R, Turley, S, Sirawaraporn, W, Hol, W.G.J. | | Deposit date: | 2000-05-05 | | Release date: | 2000-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis 7,8-dihydropteroate synthase in complex with pterin monophosphate: new insight into the enzymatic mechanism and sulfa-drug action.
J.Mol.Biol., 302, 2000
|
|
4TIM
 
 | |
4ONA
 
 | |
3TEC
 
 | | CALCIUM BINDING TO THERMITASE. CRYSTALLOGRAPHIC STUDIES OF THERMITASE AT 0, 5 AND 100 MM CALCIUM | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Kalk, K.H, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calcium binding to thermitase. Crystallographic studies of thermitase at 0, 5, and 100 mM calcium.
J.Biol.Chem., 266, 1991
|
|
4YRT
 
 | |
4YRG
 
 | |
4YRQ
 
 | |
