5H06
 
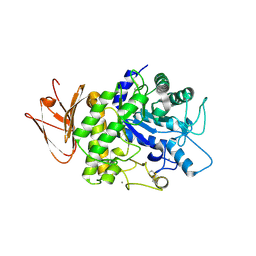 | | Crystal structure of AmyP in complex with maltose | | Descriptor: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | He, C, Liu, Y. | | Deposit date: | 2016-10-03 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
5H05
 
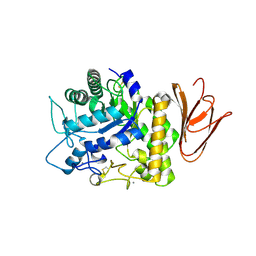 | | Crystal structure of AmyP E221Q in complex with MALTOTRIOSE | | Descriptor: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | He, C, Liu, Y. | | Deposit date: | 2016-10-03 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
3AXF
 
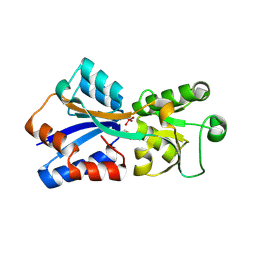 | | Perrhenate binding to A11C/R153C ModA mutant | | Descriptor: | Molybdate-binding periplasmic protein, PERRHENATE | | Authors: | He, C, Aryal, B.P, Brugarolas, P. | | Deposit date: | 2011-04-04 | | Release date: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of ReO(4) (-) with an engineered MoO (4) (2-)-binding protein: towards a new approach in radiopharmaceutical applications.
J.Biol.Inorg.Chem., 17, 2012
|
|
5HQC
 
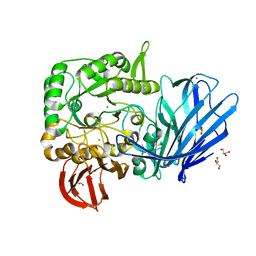 | | A Glycoside Hydrolase Family 97 enzyme R171K variant from Pseudoalteromonas sp. strain K8 | | Descriptor: | CALCIUM ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5HQB
 
 | | A Glycoside Hydrolase Family 97 enzyme (E480Q) in complex with Panose from Pseudoalteromonas sp. strain K8 | | Descriptor: | Alpha-glucosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5HQA
 
 | | A Glycoside Hydrolase Family 97 enzyme in complex with Acarbose from Pseudoalteromonas sp. strain K8 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, CALCIUM ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5HQ4
 
 | | A Glycoside Hydrolase Family 97 enzyme from Pseudoalteromonas sp. strain K8 | | Descriptor: | Alpha-glucosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5HWZ
 
 | | Crystal structure of nitrophorin 4 D30N mutant with nitrite | | Descriptor: | NITRITE ION, Nitrophorin-4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ogata, H, He, C, Lubitz, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Elucidation of the heme active site electronic structure affecting the unprecedented nitrite dismutase activity of the ferrihemebproteins, the nitrophorins.
Chem Sci, 7, 2016
|
|
3T8R
 
 | | Crystal structure of Staphylococcus aureus CymR | | Descriptor: | Staphylococcus aureus CymR | | Authors: | He, C, Ji, Q. | | Deposit date: | 2011-08-01 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Staphylococcus aureus CymR Is a New Thiol-based Oxidation-sensing Regulator of Stress Resistance and Oxidative Response.
J.Biol.Chem., 287, 2012
|
|
3TGA
 
 | |
3T8T
 
 | | Crystal structure of Staphylococcus aureus CymR oxidized form | | Descriptor: | Staphylococcus aureus CymR (oxidized form) | | Authors: | He, C, Ji, Q. | | Deposit date: | 2011-08-01 | | Release date: | 2012-05-16 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Staphylococcus aureus CymR Is a New Thiol-based Oxidation-sensing Regulator of Stress Resistance and Oxidative Response.
J.Biol.Chem., 287, 2012
|
|
3TGB
 
 | |
3TGC
 
 | |
4GNE
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-7 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GND
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains | | Descriptor: | Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GNF
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-15 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GNG
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3K9me3 peptide | | Descriptor: | GLYCEROL, Histone H3.3, Histone-lysine N-methyltransferase NSD3, ... | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4HQE
 
 | | The crystal structure of QsrR-DNA complex | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*TP*AP*AP*TP*TP*AP*TP*TP*AP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*AP*TP*AP*AP*TP*TP*AP*TP*AP*CP*T)-3'), Transcriptional regulator QsrR | | Authors: | He, C, Ji, Q, Zhang, L. | | Deposit date: | 2012-10-25 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7RFP
 
 | | Mouse GITR (mGITR) with DTA-1 Fab fragment | | Descriptor: | DTA-1 (heavy chain), DTA-1 (light chain), Tumor necrosis factor receptor superfamily member 18,Enhanced green fluorescent protein | | Authors: | Meyerson, J.R, He, C. | | Deposit date: | 2021-07-14 | | Release date: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Therapeutic antibody activation of the glucocorticoid-induced TNF receptor by a clustering mechanism.
Sci Adv, 8, 2022
|
|
7URX
 
 | | SJ25C1 Fab in complex with soluble CD19 | | Descriptor: | B-lymphocyte antigen CD19, SJ25C1 Fab heavy chain, SJ25C1 Fab light chain | | Authors: | Meyerson, J, He, C. | | Deposit date: | 2022-04-22 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CD19 CAR antigen engagement mechanisms and affinity tuning.
Sci Immunol, 8, 2023
|
|
7URV
 
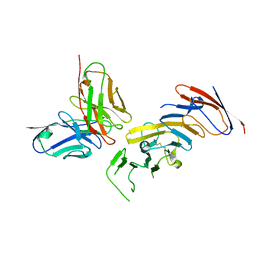 | | FMC63 scFv in complex with soluble CD19 | | Descriptor: | B-lymphocyte antigen CD19, FMC63 single-chain variable fragment | | Authors: | Meyerson, J, He, C. | | Deposit date: | 2022-04-22 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | CD19 CAR antigen engagement mechanisms and affinity tuning.
Sci Immunol, 8, 2023
|
|
4NPM
 
 | |
4NPL
 
 | |
8HKD
 
 | |
7D8M
 
 | | Crystal structure of DyP | | Descriptor: | Dye-decolorizing peroxidase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | He, C, Jia, R, Wang, T, Li, L.Q. | | Deposit date: | 2020-10-08 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revealing two important tryptophan residues with completely different roles in a dye-decolorizing peroxidase from Irpex lacteus F17.
Biotechnol Biofuels, 14, 2021
|
|
