1KCM
 
 | | Crystal Structure of Mouse PITP Alpha Void of Bound Phospholipid at 2.0 Angstroms Resolution | | Descriptor: | Phosphatidylinositol Transfer Protein alpha | | Authors: | Schouten, A, Agianian, B, Westerman, J, Kroon, J, Wirtz, K.W.A, Gros, P. | | Deposit date: | 2001-11-09 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of apo-phosphatidylinositol transfer protein alpha provides insight into membrane association.
EMBO J., 21, 2002
|
|
1M10
 
 | | Crystal structure of the complex of Glycoprotein Ib alpha and the von Willebrand Factor A1 Domain | | Descriptor: | Glycoprotein Ib alpha, von Willebrand Factor | | Authors: | Huizinga, E.G, Tsuji, S, Romijn, R.A.P, Schiphorst, M.E, de Groot, P.G, Sixma, J.J, Gros, P. | | Deposit date: | 2002-06-16 | | Release date: | 2002-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain.
Science, 297, 2002
|
|
1M0Z
 
 | | Crystal Structure of the von Willebrand Factor Binding Domain of Glycoprotein Ib alpha | | Descriptor: | Glycoprotein Ib alpha | | Authors: | Huizinga, E.G, Tsuji, S, Romijn, R.A.P, Schiphorst, M.E, de Groot, P.G, Sixma, J.J, Gros, P. | | Deposit date: | 2002-06-16 | | Release date: | 2002-08-28 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain.
Science, 297, 2002
|
|
1MNU
 
 | | UNLIGANDED BACTERICIDAL ANTIBODY AGAINST NEISSERIA MENINGITIDIS | | Descriptor: | CADMIUM ION, PROTEIN (IGG2A-KAPPA ANTIBODY MN12H2 (HEAVY CHAIN)), PROTEIN (IGG2A-KAPPA ANTIBODY MN12H2 (LIGHT CHAIN)) | | Authors: | Van Den Elsen, J, Vandeputte-Rutten, L, Kroon, J, Gros, P. | | Deposit date: | 1999-04-30 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bactericidal antibody recognition of meningococcal PorA by induced fit. Comparison of liganded and unliganded Fab structures.
J.Biol.Chem., 274, 1999
|
|
4A5W
 
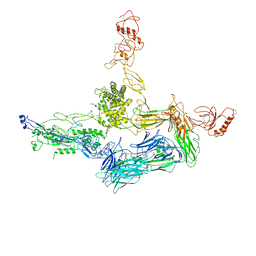 | | Crystal structure of C5b6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COMPLEMENT C5, ... | | Authors: | Hadders, M.A, Bubeck, D, Forneris, F, Pangburn, M, Llorca, O, Lea, S.M, Gros, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Assembly and Regulation of the Membrane Attack Complex Based on Structures of C5B6 and Sc5B9.
Cell Rep., 1, 2012
|
|
6FCZ
 
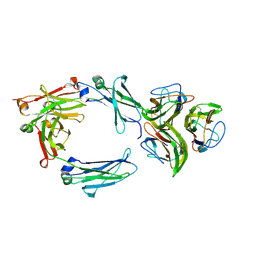 | | Model of gC1q-Fc complex based on 7A EM map | | Descriptor: | Complement C1q subcomponent subunit A, Complement C1q subcomponent subunit B, Complement C1q subcomponent subunit C, ... | | Authors: | Ugurlar, D, Howes, S.C, de Kreuk, B.J.K, de Jong, R.N, Beurskens, F.J, Koster, A.J, Parren, P.W.H.I, Sharp, T.H, Gros, P, Koning, R.I. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-28 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement.
Science, 359, 2018
|
|
6HCY
 
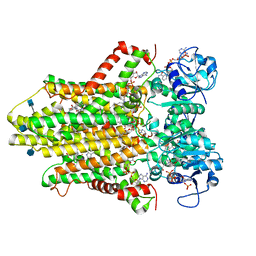 | | human STEAP4 bound to NADP, FAD, heme and Fe(III)-NTA. | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Oosterheert, W, van Bezouwen, L.S, Rodenburg, R.N.P, Forster, F, Mattevi, A, Gros, P. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human STEAP4 reveal mechanism of iron(III) reduction.
Nat Commun, 9, 2018
|
|
2BRR
 
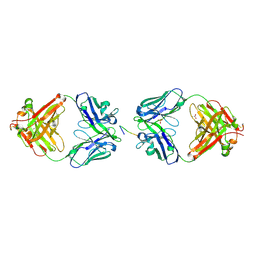 | | Complex of the neisserial PorA P1.4 epitope peptide and two Fab- fragments (antibody MN20B9.34) | | Descriptor: | ACETIC ACID, CLASS 1 OUTER MEMBRANE PROTEIN VARIABLE REGION 2, MN20B9.34 ANTI-P1.4 ANTIBODY, ... | | Authors: | Oomen, C.J, Hoogerhout, P, Kuipers, B, Vidarsson, G, Van Alphen, L, Gros, P. | | Deposit date: | 2005-05-11 | | Release date: | 2005-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Anti-Meningococcal Subtype P1.4 Pora Antibody Provides Basis for Peptide- Vaccine Design.
J.Mol.Biol., 351, 2005
|
|
2ERV
 
 | | Crystal structure of the outer membrane enzyme PagL | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL MONODECYL ETHER, hypothetical protein Paer03002360 | | Authors: | Rutten, L, Geurtsen, J, Lambert, W, Smolenaers, J.J, Bonvin, A.M, van der Ley, P, Egmond, M.R, Gros, P, Tommassen, J. | | Deposit date: | 2005-10-25 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1P4T
 
 | | Crystal structure of Neisserial surface protein A (NspA) | | Descriptor: | ETHANOLAMINE, PENTAETHYLENE GLYCOL MONODECYL ETHER, SULFATE ION, ... | | Authors: | Vandeputte-Rutten, L, Bos, M.P, Tommassen, J, Gros, P. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Neisserial Surface Protein A (NspA),
a conserved outer membrane protein with vaccine potential
J.Biol.Chem., 278, 2003
|
|
1PCF
 
 | |
1QUB
 
 | | CRYSTAL STRUCTURE OF THE GLYCOSYLATED FIVE-DOMAIN HUMAN BETA2-GLYCOPROTEIN I PURIFIED FROM BLOOD PLASMA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (human beta2-Glycoprotein I), ... | | Authors: | Bouma, B, de Groot, P.G, van den Elsen, J.M.H, Ravelli, R.B.G, Schouten, A, Simmelink, M.J.A, Derksen, R.H.W.M, Kroon, J, Gros, P. | | Deposit date: | 1999-07-01 | | Release date: | 1999-10-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Adhesion mechanism of human beta(2)-glycoprotein I to phospholipids based on its crystal structure.
EMBO J., 18, 1999
|
|
1QYN
 
 | |
2WII
 
 | | Complement C3b in complex with factor H domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COMPLEMENT C3 BETA CHAIN, ... | | Authors: | Wu, J, Janssen, B.J.C, Gros, P. | | Deposit date: | 2009-05-12 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of complement fragment C3b-factor H and implications for host protection by complement regulators.
Nat. Immunol., 10, 2009
|
|
2WIN
 
 | | C3 convertase (C3bBb) stabilized by SCIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 BETA CHAIN, ... | | Authors: | Wu, J, Janssen, B.J, Gros, P. | | Deposit date: | 2009-05-13 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor.
Nat. Immunol., 10, 2009
|
|
2XWJ
 
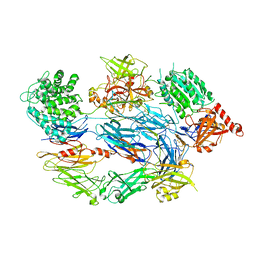 | | Crystal Structure of Complement C3b in Complex with Factor B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 ALPHA CHAIN, COMPLEMENT C3 BETA CHAIN, ... | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-04 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
2XWB
 
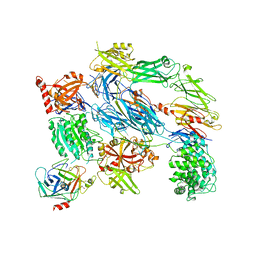 | | Crystal Structure of Complement C3b in complex with Factors B and D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3B ALPHA' CHAIN, ... | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
2XWA
 
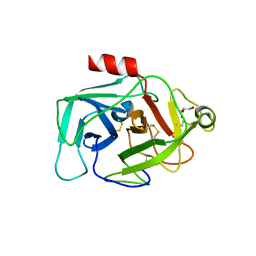 | | Crystal Structure of Complement Factor D Mutant R202A | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
2XW9
 
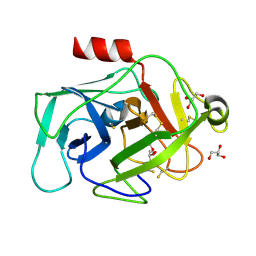 | | Crystal Structure of Complement Factor D mutant S183A | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
4AIW
 
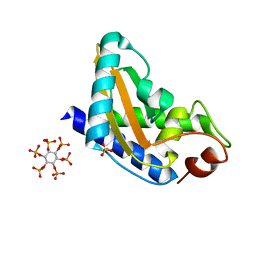 | | GAPR-1 with bound inositol hexakisphosphate | | Descriptor: | GOLGI-ASSOCIATED PLANT PATHOGENESIS-RELATED PROTEIN 1, INOSITOL HEXAKISPHOSPHATE | | Authors: | Schouten, A, Gros, P, Helms, J.B. | | Deposit date: | 2012-02-15 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interaction of Gapr-1 with Lipid Bilayers is Regulated by Alternative Homodimerization.
Biochim.Biophys.Acta, 1818, 2012
|
|
4CDK
 
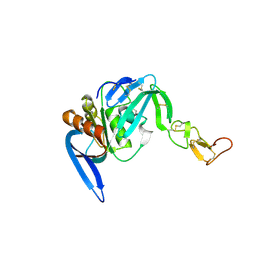 | | Structure of ZNRF3-RSPO1 | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3, R-SPONDIN-1 | | Authors: | Peng, W.C, de Lau, W, Madoori, P.K, Forneris, F, Granneman, J.C.M, Clevers, H, Gros, P. | | Deposit date: | 2013-11-01 | | Release date: | 2014-01-08 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Wnt-Antagonist Znrf3 and its Complex with R-Spondin 1 and Implications for Signaling.
Plos One, 8, 2013
|
|
4BSO
 
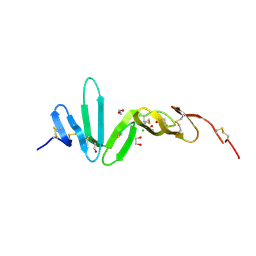 | | Crystal structure of R-spondin 1 (Fu1Fu2) - Native | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, R-SPONDIN-1 | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-19 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
4BST
 
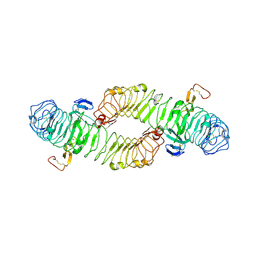 | | Structure of the ectodomain of LGR5 in complex with R-spondin-1 (Fu1Fu2) in P6122 crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING G-PROTEIN COUPLED RECEPTOR 5, R-SPONDIN-1, ... | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
4BSP
 
 | | Crystal structure of R-spondin 1 (Fu1Fu2) - Holmium soak | | Descriptor: | HOLMIUM ATOM, R-SPONDIN-1, SULFATE ION | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-19 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
4BSR
 
 | | Structure of the ectodomain of LGR5 in complex with R-spondin-1 (Fu1Fu2) in P22121 crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING G-PROTEIN COUPLED RECEPTOR 5, ... | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
