2M5B
 
 | | The NMR structure of the BID-BAK complex | | Descriptor: | Bcl-2 homologous antagonist/killer, human_BID_BH3_SAHB | | Authors: | Moldoveanu, T, Grace, C.R, Kriwacki, R.W, Green, D.R. | | Deposit date: | 2013-02-19 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | BID-induced structural changes in BAK promote apoptosis.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3T7H
 
 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
3T7G
 
 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Autophagy-related protein 3, Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
3T7F
 
 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
3T7E
 
 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
3DF0
 
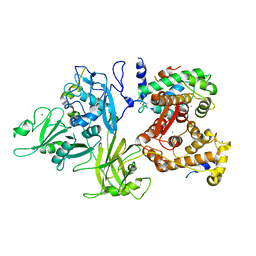 | | Calcium-dependent complex between m-calpain and calpastatin | | Descriptor: | CALCIUM ION, Calpain small subunit 1, Calpain-2 catalytic subunit, ... | | Authors: | Moldoveanu, T, Gehring, K, Green, D.R. | | Deposit date: | 2008-06-11 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains.
Nature, 456, 2008
|
|
4A1O
 
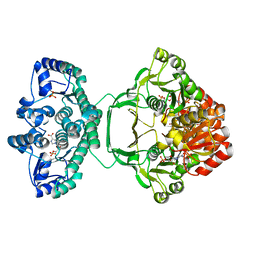 | | Crystal structure of Mycobacterium tuberculosis PurH complexed with AICAR and a novel nucleotide CFAIR, at 2.48 A resolution. | | Descriptor: | 5-(FORMYLAMINO)-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, ... | | Authors: | Le Nours, J, Bulloch, E.M.M, Zhang, Z, Greenwood, D.R, Middleditch, M.J, Dickson, J.M.J, Baker, E.N. | | Deposit date: | 2011-09-17 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide.
J.Biol.Chem., 286, 2011
|
|
3ZZM
 
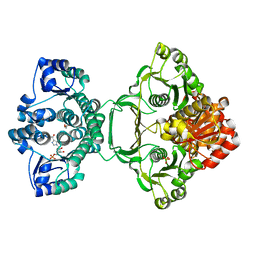 | | Crystal structure of Mycobacterium tuberculosis PurH with a novel bound nucleotide CFAIR, at 2.2 A resolution. | | Descriptor: | 5-(FORMYLAMINO)-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, GLYCEROL, ... | | Authors: | Le Nours, J, Bulloch, E.M.M, Zhang, Z, Greenwood, D.R, Middleditch, M.J, Dickson, J.M.J, Baker, E.N. | | Deposit date: | 2011-09-02 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide.
J.Biol.Chem., 286, 2011
|
|
3E8T
 
 | | Crystal Structure of Epiphyas postvittana Takeout 1 | | Descriptor: | Takeout-like protein 1, Ubiquinone-8 | | Authors: | Hamiaux, C, Stanley, D, Greenwood, D.R, Baker, E.N, Newcomb, R.D. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of Epiphyas postvittana takeout 1 with bound ubiquinone supports a role as ligand carriers for takeout proteins in insects
J.Biol.Chem., 284, 2009
|
|
3E8W
 
 | | Crystal Structure of Epiphyas postvittana Takeout 1 | | Descriptor: | Takeout-like protein 1, Ubiquinone-8 | | Authors: | Hamiaux, C, Stanley, D, Greenwood, D.R, Baker, E.N, Newcomb, R.D. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Epiphyas postvittana takeout 1 with bound ubiquinone supports a role as ligand carriers for takeout proteins in insects
J.Biol.Chem., 284, 2009
|
|
6BF2
 
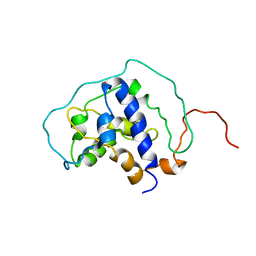 | |
6D74
 
 | |
2M04
 
 | |
2MEJ
 
 | |
2ME8
 
 | |
2ME9
 
 | |
2M03
 
 | |
4HNJ
 
 | |
4G0S
 
 | |
4C0Z
 
 | | The N-terminal domain of the Streptococcus pyogenes pilus tip adhesin Cpa | | Descriptor: | ANCILLARY PROTEIN 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Linke-Winnebeck, C, Paterson, N, Baker, E.N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-11-20 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Model for the Covalent Adhesion of the Streptococcus Pyogenes Pilus Through a Thioester Bond.
J.Biol.Chem., 289, 2014
|
|
2LHT
 
 | | Solution structure of Venturia inaequalis cellophane-induced 1 protein (ViCin1) domains 1 and 2 | | Descriptor: | Cellophane-induced protein 1 | | Authors: | Mesarich, C.H, Schmitz, M, Tremouilhac, P, Greenwood, D.R, Mcgillivray, D.J, Templeton, M.D, Dingley, A.J. | | Deposit date: | 2011-08-16 | | Release date: | 2012-07-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and domain organization of the repeat protein Cin1 from the apple scab fungus.
Biochim.Biophys.Acta, 1824, 2012
|
|
