5DD6
 
 | |
5DD3
 
 | |
5DD5
 
 | |
5F9O
 
 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235.09 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CH235.09 Light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
5F9W
 
 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of CH235-lineage antibody CH235, Light chain of CH235-lineage antibody CH235, ... | | Authors: | Chen, L, Zhou, T, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8911 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
5F96
 
 | |
4QHK
 
 | | UCA (unbound) from CH103 Lineage | | Descriptor: | UCA heavy chain, UCA light chain | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6IZ5
 
 | |
6IZ6
 
 | | Crystal Structure Analysis of TRIC counter-ion channels in calcium release | | Descriptor: | CALCIUM ION, Trimeric intracellular cation channel type B-B | | Authors: | Liu, X.L, Wang, X.H, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZF
 
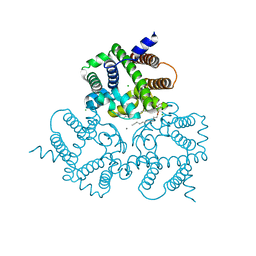 | | Structural basis for activity of TRIC counter-ion channels in calcium release | | Descriptor: | (2R)-1-(dodecanoyloxy)-3-hydroxypropan-2-yl (5E,8E,11E)-tetradeca-5,8,11-trienoate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, X.H, Zeng, Y, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZ3
 
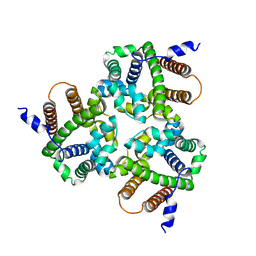 | |
8HG1
 
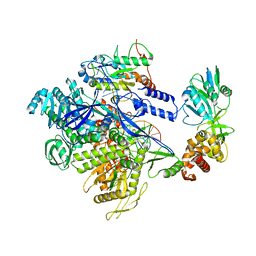 | | The structure of MPXV polymerase holoenzyme in replicating state | | Descriptor: | DNA (25-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Peng, Q, Xie, Y.F, Kuai, L, Wang, H, Qi, J.X, Gao, F, Shi, Y. | | Deposit date: | 2022-11-13 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of monkeypox virus DNA polymerase holoenzyme.
Science, 379, 2023
|
|
5U0U
 
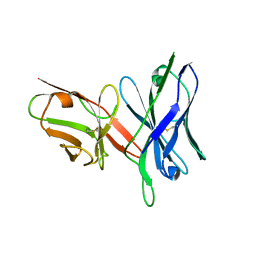 | |
5U3P
 
 | | Crystal Structure of DH511.4 Fab | | Descriptor: | DH511.4 Fab Heavy Chain, DH511.4 Fab Light Chain | | Authors: | Nicely, N.I, Williams, L.D, Ofek, G, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U15
 
 | |
5U3J
 
 | | Crystal Structure of DH511.1 Fab in Complex with HIV-1 gp41 MPER Peptide | | Descriptor: | DH511.1 Heavy Chain, DH511.1 Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5TQA
 
 | |
5U3L
 
 | | Crystal Structure of DH511.2 Fab in Complex with HIV-1 gp41 MPER 670-683 Peptide | | Descriptor: | DH511.2 Fab Heavy Chain, DH511.2 Fab Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5TRP
 
 | |
5TPP
 
 | |
5U3O
 
 | | Crystal Structure of DH511.2_K3 Fab in Complex with HIV-1 gp41 MPER Peptide | | Descriptor: | DH511.2_K3 Fab Heavy Chain, DH511.2_K3 Fab Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U3K
 
 | | Crystal Structure of DH511.2 Fab in Complex with HIV-1 gp41 MPER 662-683 Peptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, DH511.2 Fab Heavy Chain, ... | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U0R
 
 | |
5U3N
 
 | | Crystal Structure of DH511.12P Fab in Complex with HIV-1 gp41 MPER Peptide | | Descriptor: | DH511.12P Fab Heavy Chain, DH511.12P Fab Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
7YTV
 
 | |
