6CTC
 
 | |
5IDU
 
 | |
6UGK
 
 | |
4E7A
 
 | |
4E76
 
 | |
4E78
 
 | |
4EYE
 
 | |
6AMY
 
 | | Crystal structure of a 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase from Acinetobacter baumannii | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2017-08-11 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of a 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase from Acinetobacter baumannii
To Be Published
|
|
4OGX
 
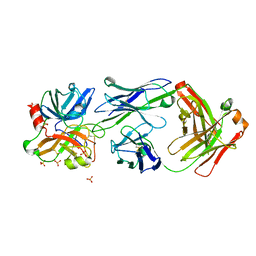 | | Crystal structure of Fab DX-2930 in complex with human plasma kallikrein at 2.4 Angstrom resolution | | Descriptor: | DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN, Plasma kallikrein, ... | | Authors: | Edwards, T.E, Clifton, M.C, Abendroth, J, Nixon, A, Ladner, R. | | Deposit date: | 2014-01-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
4OBC
 
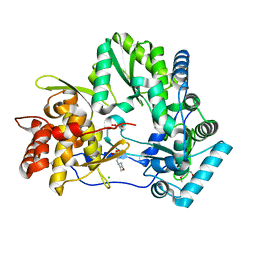 | | Crystal structure of HCV polymerase NS5b genotype 2a JFH-1 isolate with the S15G, C223H, V321I resistance mutations against the guanosine analog GS-0938 (PSI-3529238) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Edwards, T.E, Abendroth, J, Appleby, T.C. | | Deposit date: | 2014-01-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular and Structural Basis for the Roles of Hepatitis C Virus Polymerase NS5B Amino Acids 15, 223, and 321 in Viral Replication and Drug Resistance.
Antimicrob.Agents Chemother., 58, 2014
|
|
4OGY
 
 | | Crystal structure of Fab DX-2930 in complex with human plasma kallikrein at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN, ... | | Authors: | Edwards, T.E, Clifton, M.C, Abendroth, J, Nixon, A, Ladner, R. | | Deposit date: | 2014-01-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
5UJ2
 
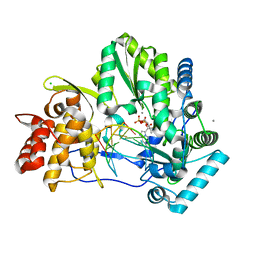 | | Crystal structure of HCV NS5B genotype 2A JFH-1 isolate with S15G E86Q E87Q C223H V321I mutations and Delta8 neta hairpoin loop deletion in complex with GS-639476 (diphsohate version of GS-9813), Mn2+ and symmetrical primer template 5'-AUAAAUUU | | Descriptor: | (1S)-1-(4-aminoimidazo[2,1-f][1,2,4]triazin-7-yl)-1,4-anhydro-2-deoxy-2-fluoro-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-2-methyl-D-ribitol, CHLORIDE ION, Genome polyprotein, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C, Murakami, E, Rey, A, McGrath, M.E. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a 2'-fluoro-2'-C-methyl C-nucleotide HCV polymerase inhibitor and a phosphoramidate prodrug with favorable properties.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5WHJ
 
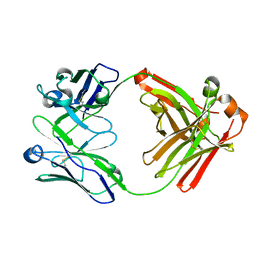 | | Crystal structure of Fab fragment of anti-FcRn antibody DX-2507 | | Descriptor: | DX-2507 Fab heavy chain, DX-2507 Fab light chain | | Authors: | Edwards, T.E, Fairman, J.W, Nixon, A.E, Kenniston, J.A. | | Deposit date: | 2017-07-17 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for pH-insensitive inhibition of immunoglobulin G recycling by an anti-neonatal Fc receptor antibody.
J. Biol. Chem., 292, 2017
|
|
5WHK
 
 | | Crystal structure of Fab fragment of antibody DX-2507 bound to FcRn-B2M | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Beta-2-microglobulin, ... | | Authors: | Edwards, T.E, Clifton, M.C, Nixon, A.E, Kenniston, J.A. | | Deposit date: | 2017-07-17 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for pH-insensitive inhibition of immunoglobulin G recycling by an anti-neonatal Fc receptor antibody.
J. Biol. Chem., 292, 2017
|
|
3IFT
 
 | | Crystal structure of glycine cleavage system protein H from Mycobacterium tuberculosis, using X-rays from the Compact Light Source. | | Descriptor: | Glycine cleavage system H protein | | Authors: | Edwards, T.E, Abendroth, J, Staker, B, Mayer, C, Phan, I, Kelley, A, Analau, E, Leibly, D, Rifkin, J, Loewen, R, Ruth, R.D, Stewart, L.J, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2009-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of the glycine cleavage system protein H of Mycobacterium tuberculosis using an inverse Compton synchrotron X-ray source.
J.Struct.Funct.Genom., 11, 2010
|
|
4WHX
 
 | | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Branched-chain-amino-acid transaminase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fairman, J.W, Dranow, D.M, Taylor, B.M, Lorimer, D, Edwards, T.E. | | Deposit date: | 2014-09-23 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate
to be published
|
|
1T4O
 
 | | Crystal structure of rnt1p dsRBD | | Descriptor: | Ribonuclease III | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, van Tilbeurgh, H, Leeper, T.C, Godin, K.S, Edwards, T.E, Sigurdsson, S.T, Rozenkrants, N, Nagel, R.J, Ares Jr, M, Varani, G. | | Deposit date: | 2004-04-30 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III
Embo J., 23, 2004
|
|
1T4N
 
 | | Solution structure of Rnt1p dsRBD | | Descriptor: | Ribonuclease III | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, van Tilbeurgh, H, Leeper, T.C, Godin, K.S, Edwards, T.E, Sigurdsson, S.T, Rozenkrants, N, Nagel, R.J, Ares, M, Varani, G. | | Deposit date: | 2004-04-30 | | Release date: | 2004-07-13 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III
Embo J., 23, 2004
|
|
7RBX
 
 | | Crystal structure of isocitrate lyase and phosphorylmutase:isocitrate lyase from Brucella melitensis biovar Abortus 2308 bound to itaconic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-methylidenebutanedioic acid, Isocitrase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Edwards, T.E, Abendroth, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aconitate decarboxylase 1 participates in the control of pulmonary Brucella infection in mice.
Plos Pathog., 17, 2021
|
|
1MX3
 
 | | Crystal structure of CtBP dehydrogenase core holo form | | Descriptor: | ACETIC ACID, C-terminal binding protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kumar, V, Carlson, J.E, Ohgi, K.E, Edwards, T.E, Rose, D.W, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2002-10-01 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transcription Corepressor CtBP Is an NAD+-Regulated Dehydrogenase
Mol.Cell, 10, 2002
|
|
8DGD
 
 | | Crystal Structure of GAF domain-containing protein, from Klebsiella pneumoniae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GAF domain-containing protein, ZINC ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Delker, S.L, Weiss, M.J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-06-23 | | Release date: | 2023-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of GAF domain-containing protein, from Klebsiella pneumoniae
to be published
|
|
5KLX
 
 | |
4QTP
 
 | | Crystal Structure of an Anti-sigma Factor Antagonist from Mycobacterium paratuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Anti-sigma factor antagonist, CITRIC ACID, ... | | Authors: | Dranow, D.M, Clifton, M.C, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of an Anti-sigma Factor Antagonist from Mycobacterium paratuberculosis
TO BE PUBLISHED
|
|
4TM5
 
 | |
4WJB
 
 | | X-ray crystal structure of a putative amidohydrolase/peptidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Putative amidohydrolase/peptidase, SULFATE ION, ... | | Authors: | Lukacs, C.M, Dranow, D.M, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of a putative amidohydrolase/peptidase from Burkholderia cenocepacia
To Be Published
|
|
