4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | Authors: | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | Deposit date: | 2015-04-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
4ZBP
 
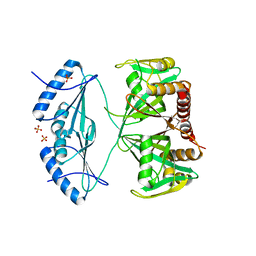 | | Crystal structure of the AMPCPR-bound AtNUDT7 | | Descriptor: | ALPHA-BETA METHYLENE ADP-RIBOSE, Nudix hydrolase 7, SULFATE ION | | Authors: | Tang, Q, Liu, C, Zhong, C, Ding, J. | | Deposit date: | 2015-04-15 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Nudix Hydrolase NUDT7 Reveal a Previously Unobserved Conformation.
Mol Plant, 8, 2015
|
|
1QK6
 
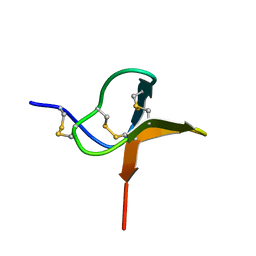 | | Solution structure of huwentoxin-I by NMR | | Descriptor: | HUWENTOXIN-I | | Authors: | Qu, Y, Liang, S, Ding, J, Liu, X, Zhang, R, Gu, X. | | Deposit date: | 1999-07-10 | | Release date: | 1999-08-20 | | Last modified: | 2019-01-16 | | Method: | SOLUTION NMR | | Cite: | Proton Nuclear Magnetic Resonance Studies on Huwentoxin-I from the Venom of the Spider Selenocosmia Huwena:2.Three-Dimensional Structure in Solution
J.Protein Chem., 16, 1997
|
|
3CXD
 
 | | Crystal structure of anti-osteopontin antibody 23C3 in complex with its epitope peptide | | Descriptor: | Fab fragment of anti-osteopontin antibody 23C3, Heavy chain, Light chain, ... | | Authors: | Du, J, Yang, H, Zhong, C, Ding, J. | | Deposit date: | 2008-04-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of recognition of human osteopontin by 23C3, a potential therapeutic antibody for treatment of rheumatoid arthritis
J.Mol.Biol., 382, 2008
|
|
3DSF
 
 | | Crystal structure of anti-osteopontin antibody 23C3 in complex with W43A mutated epitope peptide | | Descriptor: | Fab fragment of anti-osteopontin antibody 23C3, Heavy chain, Light chain, ... | | Authors: | Du, J, Zhong, C, Yang, H, Ding, J. | | Deposit date: | 2008-07-12 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of recognition of human osteopontin by 23C3, a potential therapeutic antibody for treatment of rheumatoid arthritis
J.Mol.Biol., 382, 2008
|
|
5EUE
 
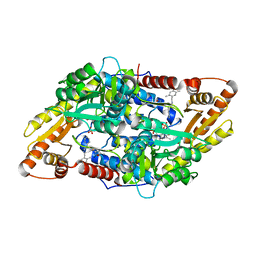 | | S1P Lyase Bacterial Surrogate bound to N-(2-((4-methoxy-2,5-dimethylbenzyl)amino)-1-phenylethyl)-5-methylisoxazole-3-carboxamide | | Descriptor: | PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ~{N}-[(1~{S})-2-[(4-methoxy-2,5-dimethyl-phenyl)methylamino]-1-phenyl-ethyl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Argiriadi, M.A, Banach, D, Radziejewska, E, Marchie, S, DiMauro, J, Dinges, J, Dominguez, E, Hutchins, C, Judge, R.A, Queeney, K, Wallace, G, Harris, C.M. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Creation of a S1P Lyase bacterial surrogate for structure-based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5EUD
 
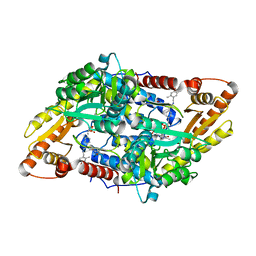 | | S1P Lyase Bacterial Surrogate bound to N-(1-(4-(3-hydroxyprop-1-yn-1-yl)phenyl)-2-((4-methoxy-2,5-dimethylbenzyl)amino)ethyl)-5-methylisoxazole-3-carboxamide | | Descriptor: | PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ~{N}-[(1~{S})-2-[(4-methoxy-2,5-dimethyl-phenyl)methylamino]-1-[4-(3-oxidanylprop-1-ynyl)phenyl]ethyl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Argiriadi, M.A, Banach, D, Radziejewska, E, Marchie, S, DiMauro, J, Dinges, J, Dominguez, E, Hutchins, C, Judge, R.A, Queeney, K, Wallace, G, Harris, C.M. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Creation of a S1P Lyase bacterial surrogate for structure-based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
2F5K
 
 | |
1O5T
 
 | | Crystal structure of the aminoacylation catalytic fragment of human tryptophanyl-tRNA synthetase | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Yu, Y, Liu, Y, Shen, N, Xu, X, Jia, J, Jin, Y, Arnold, E, Ding, J. | | Deposit date: | 2003-10-05 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Tryptophanyl-tRNA Synthetase Catalytic Fragment
J.BIOL.CHEM., 279, 2004
|
|
7D0P
 
 | |
7D0S
 
 | |
7D0Q
 
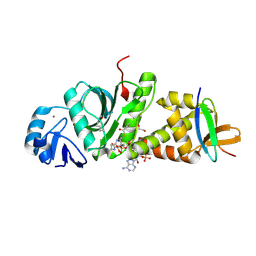 | |
7D0O
 
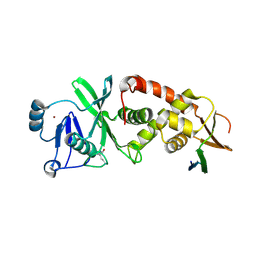 | | Crystal structure of human HBO1-BRPF2 in apo form | | Descriptor: | 1,2-ETHANEDIOL, BRD1 protein, Histone acetyltransferase KAT7, ... | | Authors: | Li, W, Ding, J. | | Deposit date: | 2020-09-11 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | HBO1 is a versatile histone acyltransferase critical for promoter histone acylations.
Nucleic Acids Res., 49, 2021
|
|
7D0R
 
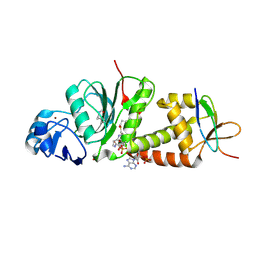 | |
3NFS
 
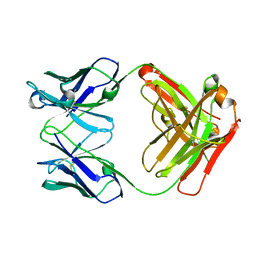 | | Crystal structure the Fab fragment of therapeutic antibody daclizumab | | Descriptor: | Heavy chain of Fab fragment of daclizumab, Light chain of Fab fragment of daclizumab | | Authors: | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
3NFP
 
 | | Crystal structure of the Fab fragment of therapeutic antibody daclizumab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of daclizumab, Interleukin-2 receptor subunit alpha, Light chain of Fab fragment of daclizumab | | Authors: | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
7F00
 
 | | Crystal structure of SPD_0310 | | Descriptor: | SULFATE ION, UPF0371 protein SPRM200_0309 | | Authors: | Cao, K, Zhang, T, Li, N, Yang, X, Ding, J, He, Q, Sun, X. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and Tetramer Structure of Hemin-Binding Protein SPD_0310 Linked to Iron Homeostasis and Virulence of Streptococcus pneumoniae.
Msystems, 7, 2022
|
|
3BKY
 
 | | Crystal Structure of Chimeric Antibody C2H7 Fab in complex with a CD20 Peptide | | Descriptor: | B-lymphocyte antigen CD20, the Fab fragment of chimeric 2H7, heavy chain, ... | | Authors: | Du, J, Zhong, C, Ding, J. | | Deposit date: | 2007-12-07 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of chimeric antibody C2H7 Fab in complex with a CD20 peptide
Mol.Immunol., 45, 2008
|
|
3OPT
 
 | | Crystal structure of the Rph1 catalytic core with a-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, DNA damage-responsive transcriptional repressor RPH1, NICKEL (II) ION | | Authors: | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
3OPW
 
 | | Crystal Structure of the Rph1 catalytic core | | Descriptor: | DNA damage-responsive transcriptional repressor RPH1 | | Authors: | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
3E9F
 
 | | Crystal structure short-form (residue1-113) of Eaf3 chromo domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromatin modification-related protein EAF3 | | Authors: | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | Deposit date: | 2008-08-22 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
3E9G
 
 | | Crystal structure long-form (residue1-124) of Eaf3 chromo domain | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | Deposit date: | 2008-08-22 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
3F6G
 
 | | Crystal structure of the regulatory domain of LiCMS in complexed with isoleucine - type II | | Descriptor: | Alpha-isopropylmalate synthase, ISOLEUCINE, SULFATE ION, ... | | Authors: | Zhang, P, Ma, J, Zhao, G, Ding, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of the inhibitor selectivity and insights into the feedback inhibition mechanism of citramalate synthase from Leptospira interrogans
Biochem.J., 421, 2009
|
|
3F6H
 
 | | Crystal structure of the regulatory domain of LiCMS in complexed with isoleucine - type III | | Descriptor: | Alpha-isopropylmalate synthase, ISOLEUCINE, ZINC ION | | Authors: | Zhang, P, Ma, J, Zhao, G, Ding, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of the inhibitor selectivity and insights into the feedback inhibition mechanism of citramalate synthase from Leptospira interrogans
Biochem.J., 421, 2009
|
|
8JDG
 
 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxybutanoic acid | | Descriptor: | (2R)-2-oxidanylbutanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-14 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
