7ROT
 
 | | Plasmodium falciparum tyrosyl-tRNA synthetase, S234C mutant, in complex with ML901-Tyr | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine--tRNA ligase, ... | | Authors: | Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy.
Science, 376, 2022
|
|
3UIL
 
 | | Crystal Structure of the complex of PGRP-S with lauric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, LAURIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-05 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3UMQ
 
 | | Crystal structure of peptidoglycan recognition protein-S complexed with butyric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, Peptidoglycan recognition protein 1, butanoic acid | | Authors: | Pandey, N, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-14 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3USX
 
 | | Crystal structure of PGRP-S complexed with Myristic Acid at 2.28 A resolution | | Descriptor: | GLYCEROL, MYRISTIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Yamini, S, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-24 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
5YSC
 
 | |
8P49
 
 | |
8QHP
 
 | |
8Q70
 
 | |
3T2V
 
 | | Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with mycolic acid at 2.5 A resolution | | Descriptor: | (2S,3R)-2-hexyl-3-hydroxynonanoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3MU7
 
 | | Crystal structure of the xylanase and alpha-amylase inhibitor protein (XAIP-II) from scadoxus multiflorus at 1.2 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, xylanase and alpha-amylase inhibitor protein | | Authors: | Kumar, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Modulation of inhibitory activity of xylanase-alpha-amylase inhibitor protein (XAIP): binding studies and crystal structure determination of XAIP-II from Scadoxus multiflorus at 1.2 A resolution.
Bmc Struct.Biol., 10, 2010
|
|
4FNN
 
 | | Crystal structure of the complex of CPGRP-S with stearic acid at 2.2 A RESOLUTION | | Descriptor: | Peptidoglycan recognition protein 1, STEARIC ACID | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site.
Arch.Biochem.Biophys., 529, 2013
|
|
3O4K
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) and lipoteichoic acid at 2.1 A resolution | | Descriptor: | (2S)-1-({3-O-[2-(acetylamino)-4-amino-2,4,6-trideoxy-beta-D-galactopyranosyl]-alpha-D-glucopyranosyl}oxy)-3-(heptanoyloxy)propan-2-yl (7Z)-pentadec-7-enoate, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis of recognition of pathogen-associated molecular patterns and inhibition of proinflammatory cytokines by camel peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
3OGX
 
 | | Crystal structure of the complex of Peptidoglycan Recognition protein (PGRP-s) with Heparin-Dissacharide at 2.8 A resolution | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of heparin binding to camel peptidoglycan recognition protein-S
Int J Biochem Mol Biol, 3, 2012
|
|
8TC8
 
 | | Human asparaginyl-tRNA synthetase bound to adenosine 5'-sulfamate | | Descriptor: | 5'-O-[N-(L-ASPARAGINYL)SULFAMOYL]ADENOSINE, Asparagine--tRNA ligase, cytoplasmic, ... | | Authors: | Dogovski, C, Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
8TC7
 
 | | Human asparaginyl-tRNA synthetase, apo form | | Descriptor: | Asparagine--tRNA ligase, cytoplasmic, CHLORIDE ION, ... | | Authors: | Dogovski, C, Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
8TC9
 
 | | Human asparaginyl-tRNA synthetase bound to OSM-S-106 | | Descriptor: | Asparagine--tRNA ligase, cytoplasmic, GLYCEROL, ... | | Authors: | Dogovski, C, Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
2W29
 
 | | Gly102Thr mutant of Rv3291c | | Descriptor: | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Shrivastava, T, Dey, S, Ravishankar, R. | | Deposit date: | 2008-10-25 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Ligand-Induced Structural Transitions, Mutational Analysis, and 'Open' Quaternary Structure of the M. Tuberculosis Feast/Famine Regulatory Protein (Rv3291C).
J.Mol.Biol., 392, 2009
|
|
3TO5
 
 | |
1Q7A
 
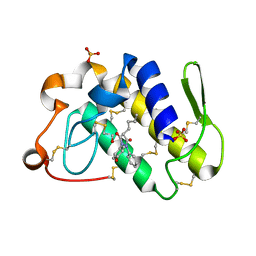 | | Crystal structure of the complex formed between russell's viper phospholipase A2 and an antiinflammatory agent oxyphenbutazone at 1.6A resolution | | Descriptor: | 4-BUTYL-1-(4-HYDROXYPHENYL)-2-PHENYLPYRAZOLIDINE-3,5-DIONE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-08-17 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phospholipase A2 as a target protein for nonsteroidal anti-inflammatory drugs (NSAIDS): crystal structure of the complex formed between phospholipase A2 and oxyphenbutazone at 1.6 A resolution.
Biochemistry, 43, 2004
|
|
1TD7
 
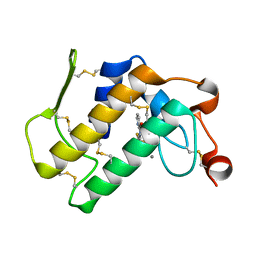 | | Interactions of a specific non-steroidal anti-inflammatory drug (NSAID) with group I phospholipase A2 (PLA2): Crystal structure of the complex formed between PLA2 and niflumic acid at 2.5 A resolution | | Descriptor: | 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Jabeen, T, Singh, N, Singh, R.K, Sharma, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-21 | | Release date: | 2004-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-steroidal anti-inflammatory drugs as potent inhibitors of phospholipase A2: structure of the complex of phospholipase A2 with niflumic acid at 2.5 Angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3NG4
 
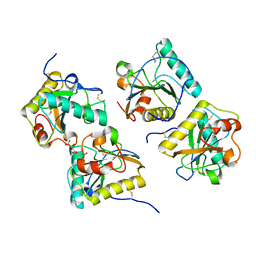 | | Ternary complex of peptidoglycan recognition protein (PGRP-S) with Maltose and N-Acetylglucosamine at 1.7 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
3NW3
 
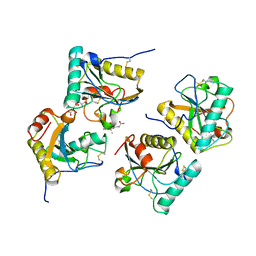 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with the PGN Fragment at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, ALANINE, D-GLUTAMINE, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
4H60
 
 | |
2B17
 
 | | Specific binding of non-steroidal anti-inflammatory drugs (NSAIDs) to phospholipase A2: Crystal structure of the complex formed between phospholipase A2 and diclofenac at 2.7 A resolution: | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2005-09-15 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Specific binding of non-steroidal anti-inflammatory drugs (NSAIDs) to phospholipase A2: structure of the complex formed between phospholipase A2 and diclofenac at 2.7 A resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4HNQ
 
 | |
