6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | Authors: | Guo, Y, Wang, S, Nie, Y, Li, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
7V6A
 
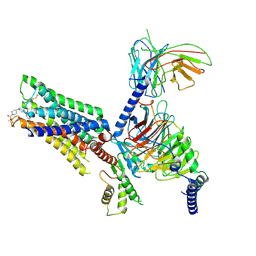 | | Cry-EM structure of M4-c110-G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.J, Wu, M, Wu, L.J, Hua, T, Liu, Z.J, Wang, T. | | Deposit date: | 2021-08-20 | | Release date: | 2022-05-11 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
7V68
 
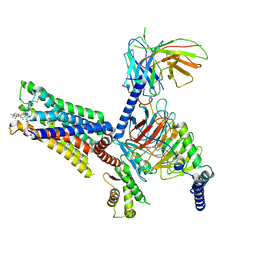 | | An Agonist and PAM-bound Class A GPCR with Gi protein complex structure | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, J.J, Wu, L.J, Wu, M, Hua, T, Liu, Z.J, Wang, T. | | Deposit date: | 2021-08-20 | | Release date: | 2022-05-11 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
7V69
 
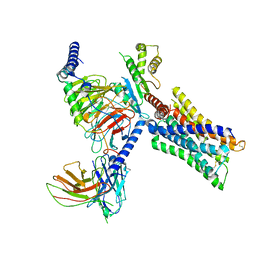 | | Cryo-EM structure of a class A GPCR-G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.J, Wu, M, Wu, L.J, Hua, T, Liu, Z.J, Wang, T. | | Deposit date: | 2021-08-20 | | Release date: | 2022-05-11 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
8T8K
 
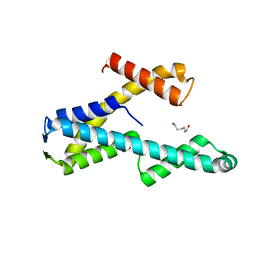 | |
8T8L
 
 | |
7YES
 
 | |
7YET
 
 | | The structure of EBOV L-VP35 in complex with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Polymerase cofactor VP35, RNA-directed RNA polymerase L | | Authors: | Shi, Y, Yuan, B, Peng, Q. | | Deposit date: | 2022-07-06 | | Release date: | 2022-10-05 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Ebola virus polymerase complex.
Nature, 610, 2022
|
|
7YER
 
 | | The structure of EBOV L-VP35 complex | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Shi, Y, Yuan, B, Peng, Q. | | Deposit date: | 2022-07-06 | | Release date: | 2022-10-05 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Ebola virus polymerase complex.
Nature, 610, 2022
|
|
7Y01
 
 | |
7PL7
 
 | |
5WPZ
 
 | | Crystal structure of MNDA PYD with MBP tag | | Descriptor: | MBP-hMNDA-PYD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
8H2F
 
 | |
8H18
 
 | |
8HI1
 
 | | Streptococcus thermophilus Cas1-Cas2- prespacer ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, DNA (26-MER), DNA (31-MER), ... | | Authors: | Chen, Q, Luo, Y. | | Deposit date: | 2022-11-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | DnaQ mediates directional spacer acquisition in the CRISPR-Cas system by a time-dependent mechanism.
Innovation (N Y), 4, 2023
|
|
8HQN
 
 | | Activation mechanism of GPR132 by 9(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQE
 
 | | Cryo-EM structure of the apo-GPR132-Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQM
 
 | | Activation mechanism of GPR132 by NPGLY | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HVI
 
 | | Activation mechanism of GPR132 by compound NOX-6-7 | | Descriptor: | 3-methyl-5-[(4-oxidanylidene-4-phenyl-butanoyl)amino]-1-benzofuran-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-26 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
6ZVI
 
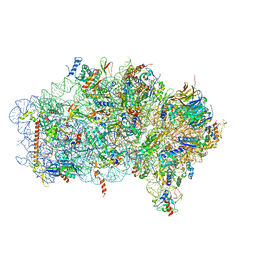 | | Mbf1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S10-A, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
6ZVH
 
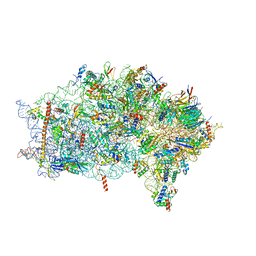 | | EDF1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
6R2M
 
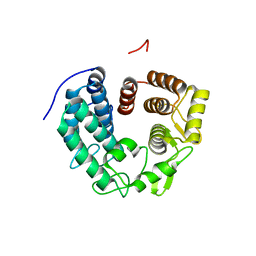 | | Crystal structure of PssZ from Listeria monocytogenes | | Descriptor: | Glycoside transferase | | Authors: | Wu, H, Cheng, J, Qiao, S, Li, D, Ma, L. | | Deposit date: | 2019-03-18 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | Crystal structure of the glycoside hydrolase PssZ from Listeria monocytogenes.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7CR6
 
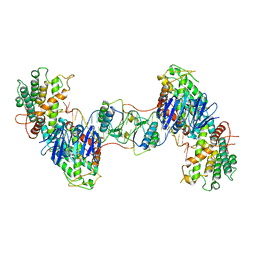 | | Synechocystis Cas1-Cas2/prespacer binary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
7CR8
 
 | | Synechocystis Cas1-Cas2-prespacerL complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
7DXJ
 
 | |
