4GEE
 
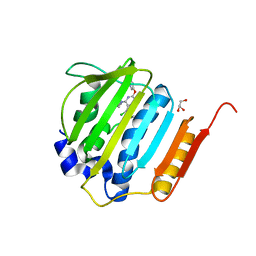 | | Pyrrolopyrimidine inhibitors of DNA gyrase B and topoisomerase IV, part I: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | (1R,5S,6s)-3-[5-chloro-6-ethyl-2-(pyrimidin-5-yloxy)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]-3-azabicyclo[3.1.0]hexan-6-amine, DNA gyrase subunit B, GLYCEROL | | Authors: | Bensen, D.C, Chen, Z, Tari, L.W. | | Deposit date: | 2012-08-01 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4FTS
 
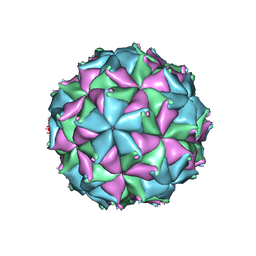 | | Crystal structure of the N363T mutant of the Flock House virus capsid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
7XV8
 
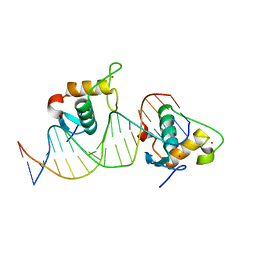 | | Crystal structure of the Human TR4 DNA-Binding Domain Homodimer Bound to DR1 Response Element | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*A)-3'), Nuclear receptor subfamily 2 group C member 2, ... | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
7XVA
 
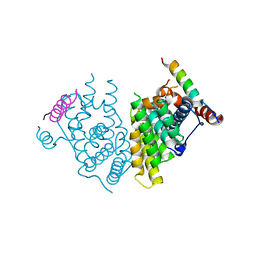 | |
7XV9
 
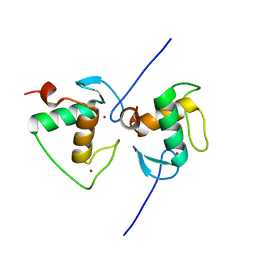 | | Crystal structure of the Human TR4 DNA-Binding Domain | | Descriptor: | Nuclear receptor subfamily 2 group C member 2, ZINC ION | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
7XV6
 
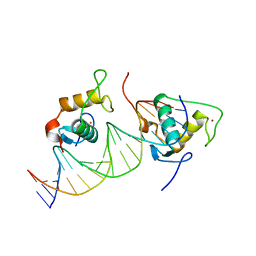 | |
4FTE
 
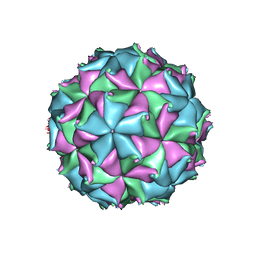 | | Crystal structure of the D75N mutant capsid of Flock House virus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
4FTB
 
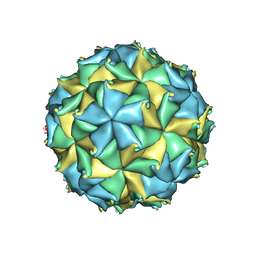 | | Crystal structure of the authentic Flock House virus particle | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
4MYW
 
 | | Structure of HSV-2 gD bound to nectin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | Authors: | Lu, G, Zhang, N, Qi, J, Li, Y, Chen, Z, Zheng, C, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.189 Å) | | Cite: | Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition.
J.Virol., 88, 2014
|
|
4MYV
 
 | | Free HSV-2 gD structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Lu, G, Zhang, N, Qi, J, Li, Y, Chen, Z, Zheng, C, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition.
J.Virol., 88, 2014
|
|
4HFP
 
 | | Structure of thrombin mutant S195a bound to the active site inhibitor argatroban | | Descriptor: | (2R,4R)-4-methyl-1-(N~2~-{[(3S)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}-L-arginyl)piperidine-2-carboxylic acid, Prothrombin, SODIUM ION | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Lin, W, Barranco-Medina, S, Pelc, L.A, Di Cera, E. | | Deposit date: | 2012-10-05 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
8HM2
 
 | |
4O03
 
 | | Crystal structure of Ca2+ bound prothrombin deletion mutant residues 146-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Shropshire, D.B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2013-12-13 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | The linker connecting the two kringles plays a key role in prothrombin activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4IAO
 
 | | Crystal structure of Sir2 C543S mutant in complex with SID domain of Sir4 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent histone deacetylase SIR2, Regulatory protein SIR4, ... | | Authors: | Hsu, H.C, Wang, C.L, Wang, M, Yang, N, Chen, Z, Sternglanz, R, Xu, R.M. | | Deposit date: | 2012-12-07 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structural basis for allosteric stimulation of Sir2 activity by Sir4 binding
Genes Dev., 27, 2013
|
|
4OBE
 
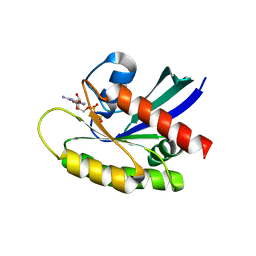 | | Crystal Structure of GDP-bound Human KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2014-01-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8HM1
 
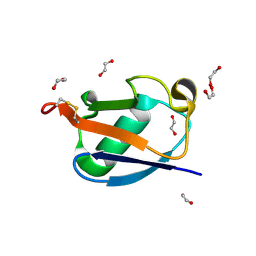 | |
8HM4
 
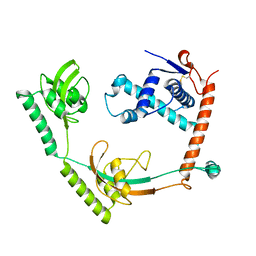 | | Crystal structure of PPIase | | Descriptor: | Peptidylprolyl isomerase | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
8HM3
 
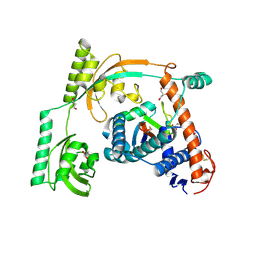 | | Complex of PPIase-BfUbb | | Descriptor: | GLYCEROL, MAGNESIUM ION, Peptidylprolyl isomerase, ... | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
8OIO
 
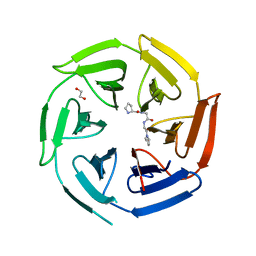 | | Crystal structure of the kelch domain of human KLHL12 in complex with PLEKHA4 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Kelch-like protein 12, ... | | Authors: | Dalietou, E.V, Chen, Z, Ramdass, A.E, Manning, C, Richardson, W, Aitmakhanova, K, Platt, M, Pike, A.C.W, Fedorov, O, Brennan, P, Bullock, A.N. | | Deposit date: | 2023-03-23 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Crystal structure of the kelch domain of human KLHL12 in complex with PLEKHA4 peptide
To Be Published
|
|
7YDW
 
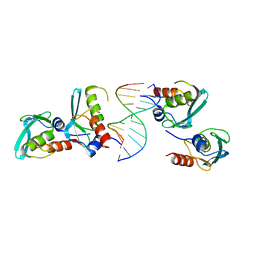 | | Crystal structure of the MPND-DNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MPN domain-containing protein | | Authors: | Yang, M, Chen, Z. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of MPND Reveal the Molecular Recognition of Nucleosomes.
Int J Mol Sci, 24, 2023
|
|
7YDT
 
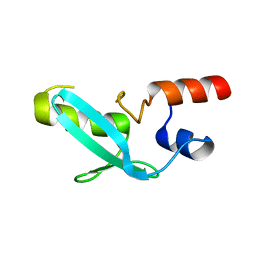 | | Crystal structure of mouse MPND | | Descriptor: | MPN domain containing protein | | Authors: | Yang, M, Chen, Z. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Structures of MPND Reveal the Molecular Recognition of Nucleosomes.
Int J Mol Sci, 24, 2023
|
|
7X3W
 
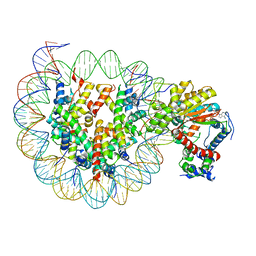 | | Cryo-EM structure of ISW1-N1 nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (146-MER), ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7X3V
 
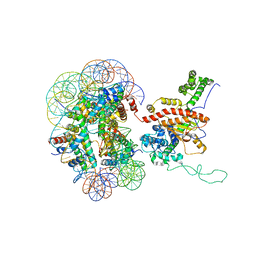 | | Cryo-EM structure of IOC3-N2 nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7X3X
 
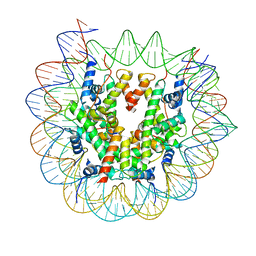 | | Cryo-EM structure of N1 nucleosome-RA | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7X3T
 
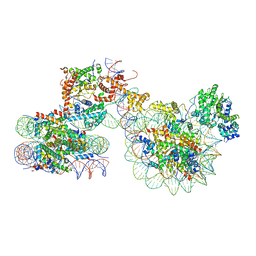 | | Cryo-EM structure of ISW1a-dinucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (343-MER), ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
