5VHO
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHM
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHP
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHF
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 10, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VGZ
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-12 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHI
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 10, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHH
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 10, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHS
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
6WIV
 
 | | Structure of human GABA(B) receptor in an inactive state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-{[(9Z)-octadec-9-enoyl]oxy}propyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Fu, Z, Frangaj, A, Liu, J, Mosyak, L, Shen, T, Slavkovich, V.N, Ray, K.M, Taura, J, Cao, B, Geng, Y, Zuo, H, Kou, Y, Grassucci, R, Chen, S, Liu, Z, Lin, X, Williams, J.P, Rice, W.J, Eng, E.T, Huang, R.K, Soni, R.K, Kloss, B, Yu, Z, Javitch, J.A, Hendrickson, W.A, Slesinger, P.A, Quick, M, Graziano, J, Yu, H, Fiehn, O, Clarke, O.B, Frank, J, Fan, Q.R. | | Deposit date: | 2020-04-10 | | Release date: | 2020-07-01 | | Last modified: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human GABABreceptor in an inactive state.
Nature, 584, 2020
|
|
4GOY
 
 | | The crystal structure of human fascin 1 K41A mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
4GP3
 
 | | The crystal structure of human fascin 1 K358A mutant | | Descriptor: | BROMIDE ION, CHLORIDE ION, Fascin, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
4GOV
 
 | | The crystal structure of human fascin 1 S39D mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
4GP0
 
 | | The crystal structure of human fascin 1 R149A K150A R151A mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
1H69
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE CO WITH 2,3,5,6,TETRAMETHYL-P-BENZOQUINONE (DUROQUINONE) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | 3-(HYDROXYMETHYL)-1-METHYL-5-(2-METHYLAZIRIDIN-1-YL)-2-PHENYL-1H-INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2001-06-08 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Development of Anticancer Drugs: Complexes of Nad(P)H:Quinone Oxidoreductase 1 with Chemotherapeutic Quinones
Structure, 9, 2001
|
|
2XCL
 
 | |
2XD4
 
 | |
4GHQ
 
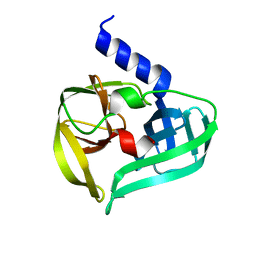 | | Crystal structure of EV71 3C proteinase | | Descriptor: | 3C proteinase | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1OZ9
 
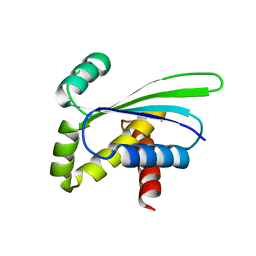 | | Crystal structure of AQ_1354, a hypothetical protein from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ_1354 | | Authors: | Oganesyan, V, Busso, D, Brandsen, J, Chen, S, Jancarik, J, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-04-08 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Structure of the hypothetical protein AQ_1354 from Aquifex aeolicus.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5C1Y
 
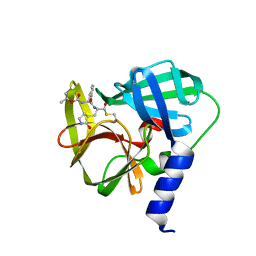 | | Crystal structure of EV71 3C Proteinase in complex with Compound 1 | | Descriptor: | 3C proteinase, propan-2-yl N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C20
 
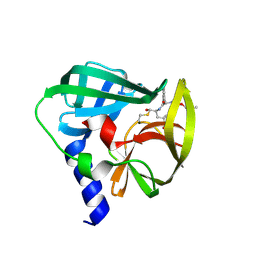 | | Crystal structure of EV71 3C Proteinase in complex with Compound 2 | | Descriptor: | 2-methylpropyl N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
4KHZ
 
 | | Crystal structure of the maltose-binding protein/maltose transporter complex in an pre-translocation conformation bound to maltoheptaose | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Binding-protein-dependent transport systems inner membrane component, Maltose transport system permease protein MalF, ... | | Authors: | Oldham, M.L, Chen, S, Chen, J. | | Deposit date: | 2013-05-01 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for substrate specificity in the Escherichia coli maltose transport system.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KI0
 
 | | Crystal structure of the maltose-binding protein/maltose transporter complex in an outward-facing conformation bound to maltohexaose | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ABC transporter related protein, Binding-protein-dependent transport systems inner membrane component, ... | | Authors: | Oldham, M.L, Chen, S, Chen, J. | | Deposit date: | 2013-05-01 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for substrate specificity in the Escherichia coli maltose transport system.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5C1U
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound Xb | | Descriptor: | (2S)-2-[[(E)-3-[4-(dimethylamino)phenyl]prop-2-enoyl]amino]-N-[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-phenyl-propanamide, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C1X
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound VIII | | Descriptor: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
