4XWY
 
 | | Crystal structure of human sepiapterin reductase in complex with an N-acetylserotinin analogue | | Descriptor: | N-[2-(5-hydroxy-2-methyl-1H-indol-3-yl)ethyl]-2-methoxyacetamide, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Johnsson, K, Hovius, R, Gorszka, K.I, Pojer, F. | | Deposit date: | 2015-01-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Reduction of Neuropathic and Inflammatory Pain through Inhibition of the Tetrahydrobiopterin Pathway.
Neuron, 86, 2015
|
|
2X6D
 
 | | Aurora-A bound to an inhibitor | | Descriptor: | 6-BROMO-7-[4-(4-CHLOROBENZYL)PIPERAZIN-1-YL]-2-[4-(MORPHOLIN-4-YLMETHYL)PHENYL]-3H-IMIDAZO[4,5-B]PYRIDINE, SERINE/THREONINE-PROTEIN KINASE 6, SULFATE ION | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Imidazo[4,5-b]pyridine derivatives as inhibitors of Aurora kinases: lead optimization studies toward the identification of an orally bioavailable preclinical development candidate.
J. Med. Chem., 53, 2010
|
|
2JEW
 
 | |
2X6E
 
 | | Aurora-A bound to an inhibitor | | Descriptor: | 6-BROMO-7-{4-[(5-METHYLISOXAZOL-3-YL)METHYL]PIPERAZIN-1-YL}-2-[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]-1H-IMIDAZO[4,5-B]PYRIDINE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Imidazo[4,5-b]pyridine derivatives as inhibitors of Aurora kinases: lead optimization studies toward the identification of an orally bioavailable preclinical development candidate.
J. Med. Chem., 53, 2010
|
|
2XOC
 
 | | C-terminal cysteine-rich domain of human CHFR bound to mADPr | | Descriptor: | ADENINE, ADENOSINE-5'-DIPHOSPHATE, E3 UBIQUITIN-PROTEIN LIGASE CHFR, ... | | Authors: | Oberoi, J, Bayliss, R. | | Deposit date: | 2010-08-11 | | Release date: | 2010-09-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis of Poly(Adp-Ribose) Recognition by the Multizinc Binding Domain of Checkpoint with Forkhead-Associated and Ring Domains (Chfr).
J.Biol.Chem., 285, 2010
|
|
2XOZ
 
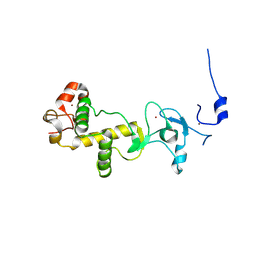 | | C-terminal cysteine rich domain of human CHFR bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, E3 UBIQUITIN-PROTEIN LIGASE CHFR, ZINC ION | | Authors: | Oberoi, J, Bayliss, R. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Structural Basis of Poly(Adp-Ribose) Recognition by the Multizinc Binding Domain of Checkpoint with Forkhead-Associated and Ring Domains (Chfr).
J.Biol.Chem., 285, 2010
|
|
2XOY
 
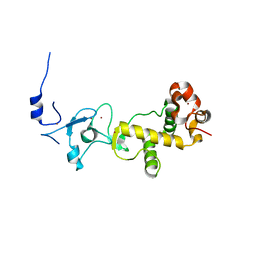 | |
2XP0
 
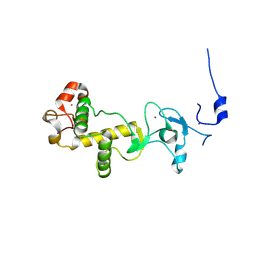 | | C-terminal cysteine-rich domain of human CHFR | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE CHFR, ZINC ION | | Authors: | Oberoi, J, Bayliss, R. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Structural Basis of Poly(Adp-Ribose) Recognition by the Multizinc Binding Domain of Checkpoint with Forkhead-Associated and Ring Domains (Chfr).
J.Biol.Chem., 285, 2010
|
|
2XNE
 
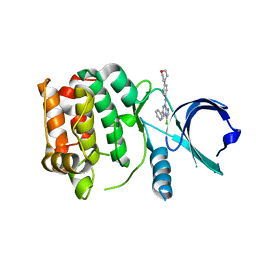 | | Structure of Aurora-A bound to an imidazopyrazine inhibitor | | Descriptor: | 3-chloro-N-(4-morpholin-4-ylphenyl)-6-pyridin-3-ylimidazo[1,2-a]pyrazin-8-amine, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
2XNG
 
 | | Structure of Aurora-A bound to a selective imidazopyrazine inhibitor | | Descriptor: | N-(3-{3-chloro-8-[(4-morpholin-4-ylphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}benzyl)methanesulfonamide, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
6H3K
 
 | | Introduction of a methyl group curbs metabolism of pyrido[3,4-d]pyrimidine MPS1 inhibitors and enables the discovery of the Phase 1 clinical candidate BOS172722. | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK, ~{N}8-(2,2-dimethylpropyl)-~{N}2-[2-ethoxy-4-(4-methyl-1,2,4-triazol-3-yl)phenyl]-6-methyl-pyrido[3,4-d]pyrimidine-2,8-diamine | | Authors: | Woodward, H.L, Hoelder, S. | | Deposit date: | 2018-07-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Introduction of a Methyl Group Curbs Metabolism of Pyrido[3,4- d]pyrimidine Monopolar Spindle 1 (MPS1) Inhibitors and Enables the Discovery of the Phase 1 Clinical Candidate N2-(2-Ethoxy-4-(4-methyl-4 H-1,2,4-triazol-3-yl)phenyl)-6-methyl- N8-neopentylpyrido[3,4- d]pyrimidine-2,8-diamine (BOS172722).
J. Med. Chem., 61, 2018
|
|
6HGT
 
 | |
2WTV
 
 | | Aurora-A Inhibitor Structure | | Descriptor: | 1,2-ETHANEDIOL, 4-{[9-CHLORO-7-(2,6-DIFLUOROPHENYL)-5H-PYRIMIDO[5,4-D][2]BENZAZEPIN-2-YL]AMINO}BENZOIC ACID, ACETATE ION, ... | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2009-09-22 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of an Aurora-A Mutant that Mimics Aurora-B Bound to Mln8054: Insights Into Selectivity and Drug Design.
Biochem.J., 427, 2010
|
|
2WTW
 
 | | Aurora-A Inhibitor Structure (2nd crystal form) | | Descriptor: | 4-{[9-CHLORO-7-(2,6-DIFLUOROPHENYL)-5H-PYRIMIDO[5,4-D][2]BENZAZEPIN-2-YL]AMINO}BENZOIC ACID, SERINE/THREONINE-PROTEIN KINASE 6 AURORA/IPL1-RELATED KINASE 1, BREAST TUMOR-AMPLIFIED KINASE, ... | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2009-09-24 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal Structure of an Aurora-A Mutant that Mimics Aurora-B Bound to Mln8054: Insights Into Selectivity and Drug Design.
Biochem.J., 427, 2010
|
|
6H1H
 
 | | Crystal structure of human Pirin in complex with compound 7 (PLX4720) | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, GLYCEROL, ... | | Authors: | Ali, S, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-11 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Privileged Structures and Polypharmacology within and between Protein Families.
ACS Med Chem Lett, 9, 2018
|
|
6H1I
 
 | | Crystal structure of human Pirin in complex with bisamide compound 2 | | Descriptor: | FE (III) ION, GLYCEROL, Pirin, ... | | Authors: | Ali, S, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-11 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Privileged Structures and Polypharmacology within and between Protein Families.
ACS Med Chem Lett, 9, 2018
|
|
5F3I
 
 | | Crystal structure of human KDM4A in complex with compound 54j | | Descriptor: | 8-[4-[2-[4-[3,5-bis(chloranyl)phenyl]piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F5C
 
 | | Crystal Structure of human JMJD2D complexed with KDOPP7 | | Descriptor: | 1,2-ETHANEDIOL, 8-[[(phenylmethyl)amino]methyl]-1~{H}-pyrido[3,4-d]pyrimidin-4-one, Lysine-specific demethylase 4D, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
4BYJ
 
 | | Aurora A kinase bound to a highly selective imidazopyridine inhibitor | | Descriptor: | (S)-N-(1-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)pyrrolidin-3-yl)acetamide, AURORA KINASE A | | Authors: | Joshi, A, Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2013-07-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Aurora Isoform Selectivity: Design and Synthesis of Imidazo[4,5-B]Pyridine Derivatives as Highly Selective Inhibitors of Aurora-A Kinase in Cells.
J.Med.Chem., 56, 2013
|
|
4BN1
 
 | | Crystal structure of V174M mutant of Aurora-A kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AURORA KINASE A, CALCIUM ION | | Authors: | Bibby, R.A, Bayliss, R. | | Deposit date: | 2013-05-13 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Insights Into Aurora-A Kinase Activation Using Unnatural Amino Acids Incorporated by Chemical Modification.
Acs Chem.Biol., 8, 2013
|
|
5F3E
 
 | | Crystal structure of human KDM4A in complex with compound 54a | | Descriptor: | 8-[4-[2-[4-(4-chlorophenyl)piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Le Bihan, Y.-V, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F37
 
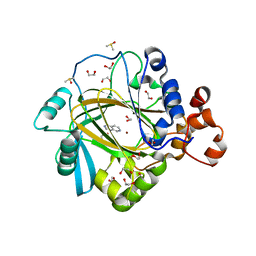 | | Crystal structure of human KDM4A in complex with compound 58 | | Descriptor: | 1,2-ETHANEDIOL, 3H-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F5A
 
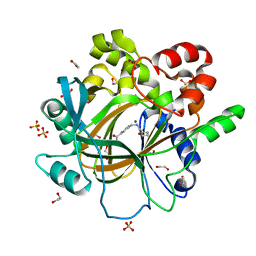 | | Crystal Structure of human JMJD2D complexed with KDOAM16 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(furan-2-ylmethylamino)methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4D, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
4BYI
 
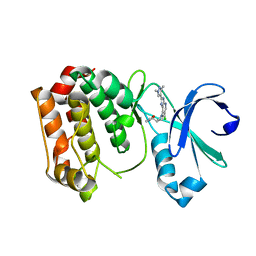 | | Aurora A kinase bound to a highly selective imidazopyridine inhibitor | | Descriptor: | (S)-N-((1-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)pyrrolidin-3-yl)methyl)acetamide, AURORA KINASE A | | Authors: | Joshi, A, Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2013-07-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aurora Isoform Selectivity: Design and Synthesis of Imidazo[4,5-B]Pyridine Derivatives as Highly Selective Inhibitors of Aurora-A Kinase in Cells.
J.Med.Chem., 56, 2013
|
|
5F2W
 
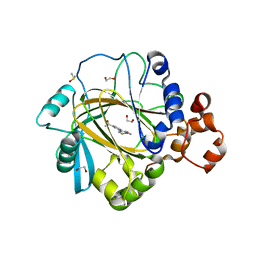 | | Crystal structure of human KDM4A in complex with compound 16 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-azanyl-1,3-thiazol-4-yl)pyridine-4-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
