2K3H
 
 | | Structural determinants for Ca2+ and PIP2 binding by the C2A domain of rabphilin-3A | | Descriptor: | CALCIUM ION, Rabphilin-3A | | Authors: | Coudevylle, N, Montaville, P, Leonov, A, Zweckstetter, M, Becker, S. | | Deposit date: | 2008-05-08 | | Release date: | 2008-10-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural Determinants for Ca2+ and Phosphatidylinositol 4,5-Bisphosphate Binding by the C2A Domain of Rabphilin-3A.
J.Biol.Chem., 283, 2008
|
|
2K39
 
 | | Recognition dynamics up to microseconds revealed from RDC derived ubiquitin ensemble in solution | | Descriptor: | Ubiquitin | | Authors: | Lange, O.F, Lakomek, N.A, Fares, C, Schroder, G, Walter, K, Becker, S, Meiler, J, Grubmuller, H, Griesinger, C, de Groot, B.L. | | Deposit date: | 2008-04-25 | | Release date: | 2008-06-24 | | Last modified: | 2019-11-13 | | Method: | SOLUTION NMR | | Cite: | Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution.
Science, 320, 2008
|
|
2KDU
 
 | | Structural basis of the Munc13-1/Ca2+-Calmodulin interaction: A novel 1-26 calmodulin binding motif with a bipartite binding mode | | Descriptor: | CALCIUM ION, Calmodulin, Protein unc-13 homolog A | | Authors: | Rodriguez-Castaneda, F.A, Maestre-Martinez, M, Coudevylle, N, Dimova, K, Jahn, O, Junge, H, Becker, S, Brose, N, Carlomagno, T, Griesinger, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-12-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Modular architecture of Munc13/calmodulin complexes: dual regulation by Ca2+ and possible function in short-term synaptic plasticity.
Embo J., 29, 2010
|
|
6V5D
 
 | | EROS3 RDC and NOE Derived Ubiquitin Ensemble | | Descriptor: | Ubiquitin | | Authors: | Lange, O.F, Lakomek, N.A, Smith, C.A, Griesinger, C, de Groot, B.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Enhancing NMR derived ensembles with kinetics on multiple timescales.
J.Biomol.Nmr, 74, 2020
|
|
4YXS
 
 | | CAMP-DEPENDENT PROTEIN KINASE PKA CATALYTIC SUBUNIT WITH PKI-5-24 | | Descriptor: | N-BENZYL-9H-PURIN-6-AMINE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Schiffer, A, Wendt, K.U. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-20 | | Last modified: | 2015-06-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A combination of spin diffusion methods for the determination of protein-ligand complex structural ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4YXR
 
 | | CRYSTAL STRUCTURE OF PKA IN COMPLEX WITH inhibitor. | | Descriptor: | 3-methyl-2H-indazole, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Schiffer, A, Wendt, K.U. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-27 | | Last modified: | 2015-06-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A combination of spin diffusion methods for the determination of protein-ligand complex structural ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7NXZ
 
 | |
6HYI
 
 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma cruzi at 1.4 A resolution in complex with inosine | | Descriptor: | INOSINE, Protein kinase A regulatory subunit | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.39944851 Å) | | Cite: | Purine nucleosides replace cAMP in allosteric regulation of PKA in trypanosomatid pathogens.
Elife, 12, 2024
|
|
4ZG4
 
 | | Myosin Vc Pre-powerstroke | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ropars, V, Pylypenko, O, Sweeney, L, Houdusse, A. | | Deposit date: | 2015-04-22 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Force-producing ADP state of myosin bound to actin.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6QCU
 
 | |
6QD8
 
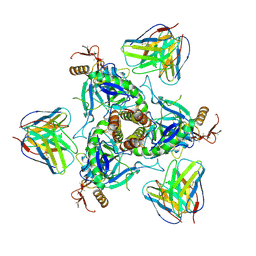 | | EM structure of a EBOV-GP bound to 4M0368 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Envelope glycoprotein,Virion spike glycoprotein,EBOV-GP1, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-01-01 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | rVSV-ZEBOV induces a polyclonal and convergent B cell response with potent Ebola virus-neutralizing antibodies
Nat.Med. (N.Y.), 2019
|
|
7QI2
 
 | |
1H2C
 
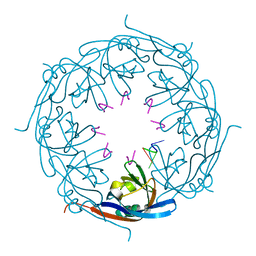 | | Ebola virus matrix protein VP40 N-terminal domain in complex with RNA (High-resolution VP40[55-194] variant). | | Descriptor: | 5'-R(*UP*GP*AP)-3', MATRIX PROTEIN VP40 | | Authors: | Gomis-Ruth, F.X, Dessen, A, Bracher, A, Klenk, H.D, Weissenhorn, W. | | Deposit date: | 2002-08-05 | | Release date: | 2003-04-10 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Matrix Protein Vp40 from Ebola Virus Octamerizes Into Pore-Like Structures with Specific RNA Binding Properties
Structure, 11, 2003
|
|
1H2D
 
 | | Ebola virus matrix protein VP40 N-terminal domain in complex with RNA (Low-resolution VP40[31-212] variant). | | Descriptor: | 5'-R(*UP*GP*AP)-3', CHLORIDE ION, MATRIX PROTEIN VP40 | | Authors: | Gomis-Ruth, F.X, Dessen, A, Bracher, A, Klenk, H.D, Weissenhorn, W. | | Deposit date: | 2002-08-06 | | Release date: | 2003-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Matrix Protein Vp40 from Ebola Virus Octamerizes Into Pore-Like Structures with Specific RNA Binding Properties
Structure, 11, 2003
|
|
6QD7
 
 | | EM structure of a EBOV-GP bound to 3T0331 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Envelope glycoprotein,Virion spike glycoprotein,EBOV-GP1, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-01-01 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV.
Nat. Med., 25, 2019
|
|
6R99
 
 | |
2LGC
 
 | |
6Y2G
 
 | | Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
6Y2F
 
 | | Crystal structure (monoclinic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
6Y7M
 
 | | Crystal structure of the complex resulting from the reaction between the SARS-CoV main protease and tert-butyl (1-((S)-3-cyclohexyl-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclohexyl-1-[[(2~{S},3~{R})-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Hilgenfeld, R. | | Deposit date: | 2020-03-01 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
6Y2E
 
 | |
6FLO
 
 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma brucei at 2.1 Angstrom resolution | | Descriptor: | GLYCEROL, INOSINE, Protein kinase A regulatory subunit | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-01-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.13868666 Å) | | Cite: | Purine nucleosides replace cAMP in allosteric regulation of PKA in trypanosomatid pathogens.
Elife, 12, 2024
|
|
6H4G
 
 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma brucei: E311A, T318R, V319A mutant bound to cAMP in the A site | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, INOSINE, Protein kinase A regulatory subunit | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-07-21 | | Release date: | 2019-08-14 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.143517 Å) | | Cite: | Purine nucleosides replace cAMP in allosteric regulation of PKA in trypanosomatid pathogens.
Elife, 12, 2024
|
|
1GIB
 
 | | MU-CONOTOXIN GIIIB, NMR | | Descriptor: | MU-CONOTOXIN GIIIB | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-04-17 | | Release date: | 1996-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mu-conotoxin GIIIB, a specific blocker of skeletal muscle sodium channels.
Biochemistry, 35, 1996
|
|
7JZJ
 
 | |
