7KUI
 
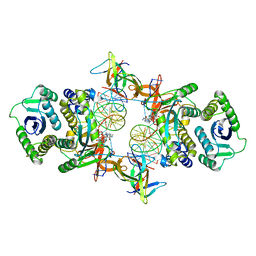 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. CIC region of a cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-25 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
7KU7
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. Cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
7M8R
 
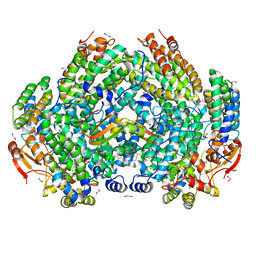 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit with fluorosubstituted tryptophans | | Descriptor: | 1,1,1-tris(fluoranyl)propan-2-one, 1,2-ETHANEDIOL, BENZOIC ACID, ... | | Authors: | Johns, J.C, Banerjee, R, Shi, K, Semonis, M.M, Aihara, H, Pomerantz, W.C.K, Lipscomb, J.D. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Soluble Methane Monooxygenase Component Interactions Monitored by 19 F NMR.
Biochemistry, 60, 2021
|
|
7M8Q
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit with fluorosubstituted tryptophans | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Shi, K, Semonis, M.M, Aihara, H, Pomerantz, W.C.K, Lipscomb, J.D. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Soluble Methane Monooxygenase Component Interactions Monitored by 19 F NMR.
Biochemistry, 60, 2021
|
|
7LRO
 
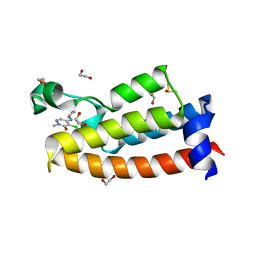 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-01-105 | | Descriptor: | 1,2-ETHANEDIOL, 5-(azetidin-3-ylamino)-4-chloranyl-2-methyl-pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LRK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-093 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{S})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LP0
 
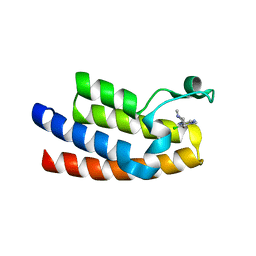 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-077 | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorol-2-methyl-5-[[(3~{R})-1-methylpiperidin-3-yl]amino]pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7M2E
 
 | | Crystal structure of BPTF bromodomain in complex with CB02-092 | | Descriptor: | 4-chloro-5-{4-[2-(dimethylamino)ethyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Nithianantham, S, Fischer, M. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition
J.Med.Chem., 64, 2021
|
|
8FZC
 
 | |
2NBW
 
 | | Solution structure of the Rpn1 T1 site with the Rad23 UBL domain | | Descriptor: | 26S proteasome regulatory subunit RPN1, UV excision repair protein RAD23 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2016-03-14 | | Release date: | 2016-07-20 | | Last modified: | 2016-08-17 | | Method: | SOLUTION NMR | | Cite: | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
2NBV
 
 | |
2NBU
 
 | |
6Q1M
 
 | | Crystal structure of the wheat dwarf virus Rep domain | | Descriptor: | GLYCEROL, Replication-associated protein | | Authors: | Litzau, L.A, Everett, B.A, Evans III, R.L, Shi, K, Tompkins, K, Gordon, W.R. | | Deposit date: | 2019-08-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of the Wheat dwarf virus Rep domain.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6WDZ
 
 | | Porcine circovirus 2 Rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent helicase Rep, DNA (5'-D(*TP*AP*GP*TP*AP*TP*TP*AP*CP*C)-3'), ... | | Authors: | Litzau, L.A, Tompkins, K, Shi, K, Nelson, A, Evans III, R.L, Gordon, W.R. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering.
Nucleic Acids Res., 49, 2021
|
|
6WE1
 
 | | Wheat dwarf virus Rep domain complexed with a single-stranded DNA 8-mer comprising the cleavage site | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*TP*TP*AP*C)-3'), MANGANESE (II) ION, Replication-associated protein | | Authors: | Tompkins, K, Litzau, L.A, Pornschloegl, L, Nelson, A.T, Evans III, R.L, Gordon, W.R. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering.
Nucleic Acids Res., 49, 2021
|
|
6WE0
 
 | | Wheat dwarf virus Rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*AP*TP*TP*AP*CP*C)-3'), MANGANESE (II) ION, Replication-associated protein | | Authors: | Tompkins, K, Litzau, L.A, Shi, K, Nelson, A, Evans III, R.L, Gordon, W.R. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering.
Nucleic Acids Res., 49, 2021
|
|
6CA4
 
 | | Crystal structure of humanized D. rerio TDP2 by 14 mutations | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Shi, K, Aihrara, H. | | Deposit date: | 2018-01-29 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | New fluorescence-based high-throughput screening assay for small molecule inhibitors of tyrosyl-DNA phosphodiesterase 2 (TDP2).
Eur J Pharm Sci, 118, 2018
|
|
7LPK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-03-112 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{R})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-12 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
8SNI
 
 | | Hydroxynitrile Lyase from Hevea brasiliensis with Forty Mutations | | Descriptor: | (S)-hydroxynitrile lyase, 1,2-ETHANEDIOL, BENZOIC ACID, ... | | Authors: | Walsh, M.E, Greenberg, L.R, Kazlauskas, R.J, Pierce, C.T, Aihara, H, Evans, R.L, Shi, K. | | Deposit date: | 2023-04-27 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | To be published
To Be Published
|
|
