6M4F
 
 | | Crystal structure of the E496A mutant of HsBglA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase-like enzyme, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
1IDZ
 
 | |
1IDY
 
 | |
1CK6
 
 | | BINDING MODE OF SALICYLHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN (PEROXIDASE), ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1999-04-28 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of salicylhydroxamic acid and several aromatic donor molecules to Arthromyces ramosus peroxidase, investigated by X-ray crystallography, optical difference spectroscopy, NMR relaxation, molecular dynamics, and kinetics.
Biochemistry, 38, 1999
|
|
5AY8
 
 | | Crystal structure of human nucleosome containing H3.Y | | Descriptor: | CHLORIDE ION, DNA (146-MER), H3.Y, ... | | Authors: | Kujirai, T, Horikoshi, N, Sato, K, Maehara, K, Machida, S, Osakabe, A, Kimura, H, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-08-10 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of human histone H3.Y nucleosome
Nucleic Acids Res., 44, 2016
|
|
5B13
 
 | | Crystal structure of phycoerythrin | | Descriptor: | PHYCOCYANOBILIN, PHYCOUROBILIN, Phycoerythrin alpha subunit, ... | | Authors: | Tanaka, Y, Gai, Z, Kishimura, H. | | Deposit date: | 2015-11-18 | | Release date: | 2016-10-05 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Structural properties of phycoerythrin from dulse palmaria palmata
J FOOD BIOCHEM., 2016
|
|
5B37
 
 | | Crystal structure of L-tryptophan dehydrogenase from Nostoc punctiforme | | Descriptor: | Tryptophan dehydrogenase | | Authors: | Wakamatsu, T, Sakuraba, H, Kitamura, M, Hakumai, Y, Ohnishi, K, Ashiuchi, M, Ohshima, T. | | Deposit date: | 2016-02-11 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Insights into l-Tryptophan Dehydrogenase from a Photoautotrophic Cyanobacterium, Nostoc punctiforme.
Appl. Environ. Microbiol., 83, 2017
|
|
1GOA
 
 | | COOPERATIVE STABILIZATION OF ESCHERICHIA COLI RIBONUCLEASE HI BY INSERTION OF GLY-80B AND GLY-77-> ALA SUBSTITUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Nakamura, H, Morikawa, K, Kanaya, S. | | Deposit date: | 1993-05-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cooperative stabilization of Escherichia coli ribonuclease HI by insertion of Gly-80b and Gly-77-->Ala substitution.
Biochemistry, 32, 1993
|
|
1GOC
 
 | | COOPERATIVE STABILIZATION OF ESCHERICHIA COLI RIBONUCLEASE HI BY INSERTION OF GLY-80B AND GLY-77-> ALA SUBSTITUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Nakamura, H, Morikawa, K, Kanaya, S. | | Deposit date: | 1993-05-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperative stabilization of Escherichia coli ribonuclease HI by insertion of Gly-80b and Gly-77-->Ala substitution.
Biochemistry, 32, 1993
|
|
4YN8
 
 | | Crystal Structure of Response Regulator ChrA in Heme-Sensing Two Component System | | Descriptor: | MAGNESIUM ION, Response regulator ChrA, SULFATE ION | | Authors: | Doi, A, Nakamura, H, Shiro, Y, Sugimoto, H. | | Deposit date: | 2015-03-09 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the response regulator ChrA in the haem-sensing two-component system of Corynebacterium diphtheriae.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1GOB
 
 | | COOPERATIVE STABILIZATION OF ESCHERICHIA COLI RIBONUCLEASE HI BY INSERTION OF GLY-80B AND GLY-77-> ALA SUBSTITUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Nakamura, H, Morikawa, K, Kanaya, S. | | Deposit date: | 1993-05-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperative stabilization of Escherichia coli ribonuclease HI by insertion of Gly-80b and Gly-77-->Ala substitution.
Biochemistry, 32, 1993
|
|
1IOX
 
 | | NMR Structure of human Betacellulin-2 | | Descriptor: | Betacellulin | | Authors: | Miura, K, Doura, H, Aizawa, T, Tada, H, Seno, M, Yamada, H, Kawano, K. | | Deposit date: | 2001-04-18 | | Release date: | 2002-07-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of betacellulin, a new member of EGF-family ligands.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
1D06
 
 | | STRUCTURAL BASIS OF DIMERIZATION AND SENSORY MECHANISMS OF OXYGEN-SENSING DOMAIN OF RHIZOBIUM MELILOTI FIXL DETERMINED AT 1.4A RESOLUTION | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, nitrogen fixation regulatory protein fixL | | Authors: | Miyatake, H, Mukai, M, Park, S.-Y, Adachi, S, Tamura, K, Nakamura, H, Nakamura, K, Tsuchiya, T, Iizuka, T, Shiro, Y. | | Deposit date: | 1999-09-09 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies
J.MOL.BIOL., 301, 2000
|
|
1JIQ
 
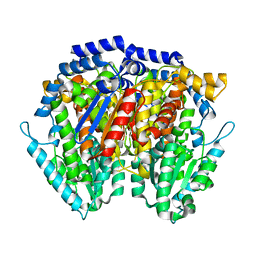 | | Crystal Structure of Human Autocrine Motility Factor | | Descriptor: | autocrine motility factor | | Authors: | Tanaka, N, Haga, A, Uemura, H, Akiyama, H, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2001-07-02 | | Release date: | 2002-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition mechanism of cytokine activity of human autocrine motility factor examined by crystal structure analyses and site-directed mutagenesis studies.
J.Mol.Biol., 318, 2002
|
|
1IRI
 
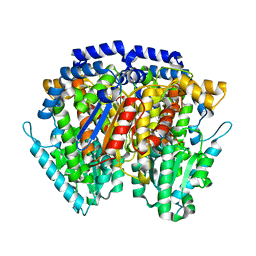 | | Crystal structure of human autocrine motility factor complexed with an inhibitor | | Descriptor: | ERYTHOSE-4-PHOSPHATE, autocrine motility factor | | Authors: | Tanaka, N, Haga, A, Uemura, H, Akiyama, H, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2001-10-08 | | Release date: | 2002-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition mechanism of cytokine activity of human autocrine motility factor examined by crystal structure analyses and site-directed mutagenesis studies.
J.Mol.Biol., 318, 2002
|
|
1V40
 
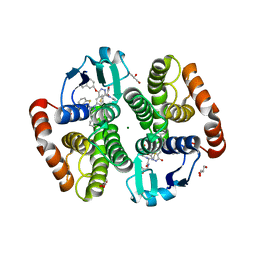 | | First Inhibitor Complex Structure of Human Hematopoietic Prostaglandin D Synthase | | Descriptor: | 3-(1,3-BENZOTHIAZOL-2-YL)-2-(1,4-DIOXO-1,2,3,4-TETRAHYDROPHTHALAZIN-6-YL)-5-[(E)-2-PHENYLVINYL]-3H-TETRAAZOL-2-IUM, GLUTATHIONE, GLYCEROL, ... | | Authors: | Inoue, T, Okano, Y, Kado, Y, Aritake, K, Irikura, D, Uodome, N, Kinugasa, S, Okazaki, N, Matsumura, H, Kai, Y, Urade, Y. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First determination of the inhibitor complex structure of human hematopoietic prostaglandin D synthase.
J.Biochem.(Tokyo), 135, 2004
|
|
1IP0
 
 | | NMR STRUCTURE OF HUMAN BETACELLULIN-2 | | Descriptor: | BETACELLULIN | | Authors: | Miura, K, Doura, H, Aizawa, T, Tada, H, Seno, M, Yamada, H, Kawano, K. | | Deposit date: | 2001-04-19 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of betacellulin, a new member of EGF-family ligands.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
6JB2
 
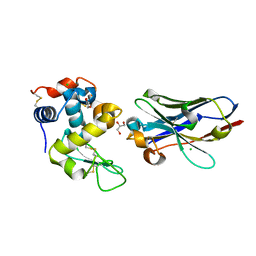 | | Crystal structure of nanobody D3-L11 mutant Y102A in complex with hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
6JB9
 
 | | Crystal structure of nanobody D3-L11 (unbound form) | | Descriptor: | Nanobody D3-L11, SULFATE ION | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
8J81
 
 | | MDM2 bound with a peptoid | | Descriptor: | (2S)-2-[[(2S)-2-[(6-chloranyl-1H-indol-3-yl)methyl-[(2S)-2-[[(2S)-2-[ethanoyl-(phenylmethyl)amino]propanoyl]-methyl-amino]propanoyl]amino]propanoyl]-methyl-amino]-N-(3,3-dimethylbutyl)-N-[(2S)-1-oxidanylidene-1-piperazin-1-yl-propan-2-yl]propanamide, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Yokomine, M, Fukuda, Y, Ago, H, Matsuura, H, Ueno, G, Nagatoishi, S, Yamamoto, M, Tsumoto, K, Jumpei, M, Sando, S. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A structural and physicochemical study of how a peptoid binds to a protein
To Be Published
|
|
6J8Y
 
 | | Crystal structure of the human RAD9-HUS1-RAD1-RHINO complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Iida, N, Sakurai, H, Hashimoto, H. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAD9-RAD1-HUS1 checkpoint clamp bound to RHINO sheds light on the other side of the DNA clamp.
J.Biol.Chem., 295, 2020
|
|
6JB8
 
 | | Crystal structure of nanobody D3-L11 in complex with hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
1WPG
 
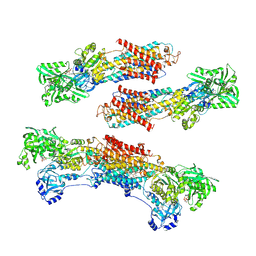 | | Crystal structure of the SR CA2+-ATPase with MGF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Toyoshima, C, Nomura, H, Tsuda, T. | | Deposit date: | 2004-09-02 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues
Nature, 432, 2004
|
|
1MBJ
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
