8WAT
 
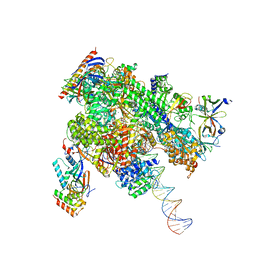 | | De novo transcribing complex 10 (TC10), the early elongation complex with Pol II positioned 10nt downstream of TSS | | Descriptor: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WB0
 
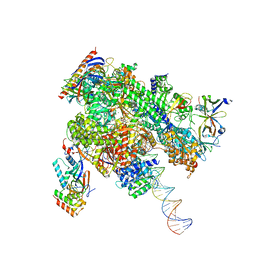 | | De novo transcribing complex 17 (TC17), the early elongation complex with Pol II positioned 17nt downstream of TSS | | Descriptor: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAU
 
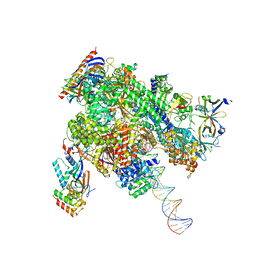 | | De novo transcribing complex 11 (TC11), the early elongation complex with Pol II positioned 11nt downstream of TSS | | Descriptor: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
6Q06
 
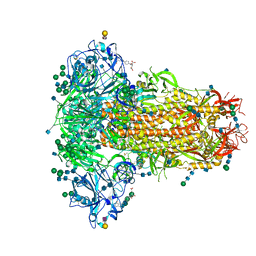 | | MERS-CoV S structure in complex with 2,3-sialyl-N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q05
 
 | | MERS-CoV S structure in complex with sialyl-lewisX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6AGK
 
 | | The structure of CH-II-77-tubulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, H, Arnst, K, Wang, Y, Miller, D, Li, W. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationship Study of Novel 6-Aryl-2-benzoyl-pyridines as Tubulin Polymerization Inhibitors with Potent Antiproliferative Properties.
J.Med.Chem., 63, 2020
|
|
8TGN
 
 | | VMAT1 dimer with serotonin and reserpine | | Descriptor: | Chromaffin granule amine transporter, SEROTONIN, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGL
 
 | | VMAT1 dimer with norepinephrine and reserpine | | Descriptor: | Chromaffin granule amine transporter, L-NOREPINEPHRINE, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGI
 
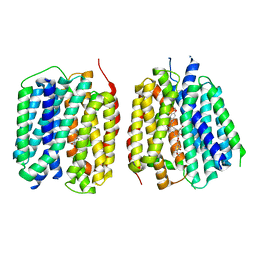 | | VMAT1 dimer with dopamine and reserpine | | Descriptor: | Chromaffin granule amine transporter, L-DOPAMINE, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGM
 
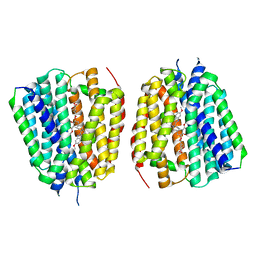 | | VMAT1 dimer with reserpine | | Descriptor: | VMAT1 dimer with reserpine, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGG
 
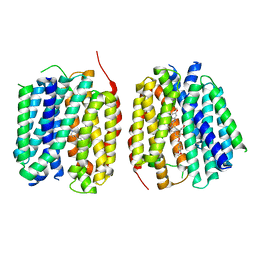 | | VMAT1 dimer with MPP+ and reserpine | | Descriptor: | 1-methyl-4-phenylpyridin-1-ium, Chromaffin granule amine transporter, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGH
 
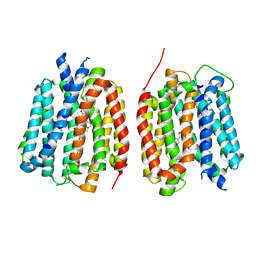 | | VMAT1 dimer with amphetamine and reserpine | | Descriptor: | (2S)-1-phenylpropan-2-amine, Chromaffin granule amine transporter, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGK
 
 | | VMAT1 dimer with histamine and reserpine | | Descriptor: | Chromaffin granule amine transporter, HISTAMINE, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGJ
 
 | | VMAT1 dimer in unbound form and with reserpine | | Descriptor: | VMAT1 dimer in unbound form and with reserpine, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
6O7K
 
 | | 30S initiation complex | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Frank, J, Gonzalez Jr, R.L, kaledhonkar, S, Fu, Z, Caban, K, Li, W, Chen, B, Sun, M. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Late steps in bacterial translation initiation visualized using time-resolved cryo-EM.
Nature, 570, 2019
|
|
6Q07
 
 | | MERS-CoV S structure in complex with 2,6-sialyl-N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2P3V
 
 | | Thermotoga maritima IMPase TM1415 | | Descriptor: | Inositol-1-monophosphatase, S,R MESO-TARTARIC ACID | | Authors: | Stieglitz, K.A, Roberts, M.F, Li, W, Stec, B. | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the tetrameric inositol 1-phosphate phosphatase (TM1415) from the hyperthermophile, Thermotoga maritima.
Febs J., 274, 2007
|
|
8KD5
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD4
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KC7
 
 | | Rpd3S histone deacetylase complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-06 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD7
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA | | Descriptor: | 167bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
1G7P
 
 | | CRYSTAL STRUCTURE OF MHC CLASS I H-2KB HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND YEAST ALPHA-GLUCOSIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-GLUCOSIDASE P1, ... | | Authors: | Apostolopoulos, V, Yu, M, Corper, A.L, Li, W, McKenzie, I.F, Teyton, L, Wilson, I.A. | | Deposit date: | 2000-11-13 | | Release date: | 2002-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a non-canonical high affinity peptide complexed with MHC class I: a novel use of alternative anchors.
J.Mol.Biol., 318, 2002
|
|
