4UI1
 
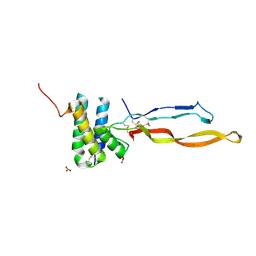 | | Crystal structure of the human RGMC-BMP2 complex | | Descriptor: | 1,2-ETHANEDIOL, BONE MORPHOGENETIC PROTEIN 2, CHLORIDE ION, ... | | Authors: | Healey, E.G, Bishop, B, Elegheert, J, Bell, C.H, Padilla-Parra, S, Siebold, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Repulsive Guidance Molecule is a Structural Bridge between Neogenin and Bone Morphogenetic Protein.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UIV
 
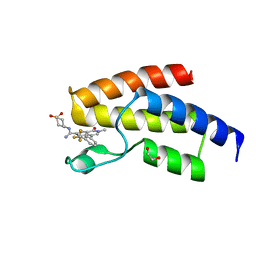 | | BROMODOMAIN OF HUMAN BRD9 WITH N-(1,1-dioxo-1-thian-4-yl)-5-methyl-4- oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2- carboximidamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 9, N-(1,1-dioxo-1-thian-4-yl)-5-methyl-4-oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2-carboximidamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
4U79
 
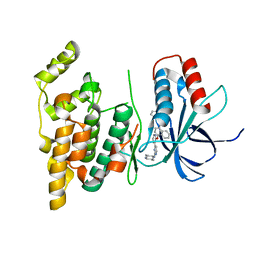 | | Crystal structure of human JNK3 in complex with a benzenesulfonamide inhibitor. | | Descriptor: | Mitogen-activated protein kinase 10, N-{4-[(3-{2-[(trans-4-aminocyclohexyl)amino]pyrimidin-4-yl}pyridin-2-yl)oxy]naphthalen-1-yl}benzenesulfonamide | | Authors: | Mohr, C. | | Deposit date: | 2014-07-30 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Unfolded Protein Response in Cancer: IRE1 alpha Inhibition by Selective Kinase Ligands Does Not Impair Tumor Cell Viability.
Acs Med.Chem.Lett., 6, 2015
|
|
8Q1O
 
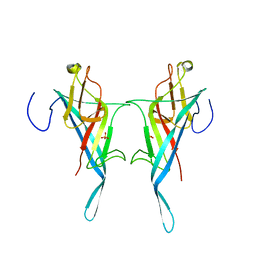 | | S-layer protein SlpA from Lactobacillus amylovorus, domain I (aa 32-209), important for Self-assembly | | Descriptor: | PHOSPHATE ION, S-layer | | Authors: | Sagmeister, T, Grininger, C, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2023-08-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4UDE
 
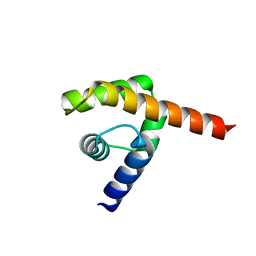 | | An oligomerization domain confers pioneer properties to the LEAFY master floral regulator | | Descriptor: | GINLFY PROTEIN, GLYCEROL, TETRAETHYLENE GLYCOL | | Authors: | Nanao, M.H, Sayou, C, Dumas, R, Parcy, F. | | Deposit date: | 2014-12-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Sam Oligomerization Domain Shapes the Genomic Binding Landscape of the Leafy Transcription Factor
Nat.Commun., 7, 2016
|
|
4UCW
 
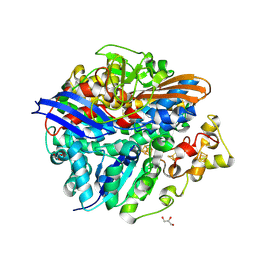 | | Structure of the T18V small subunit mutant of D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Abou-Hamdan, A, Ceccaldi, P, Lebrette, H, Guttierez-Sanz, O, Richaud, P, Cournac, L, Guigliarelli, B, deLacey, A.L, Leger, C, Volbeda, A, Burlat, B, Dementin, S. | | Deposit date: | 2014-12-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Threonine Stabilizes the Nic and Nir Catalytic Intermediates of [Nife]-Hydrogenase.
J.Biol.Chem., 290, 2015
|
|
4UF8
 
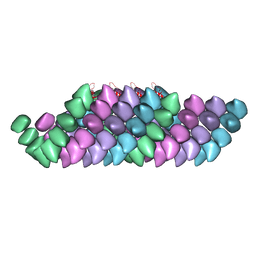 | | Electron cryo-microscopy structure of PB1-p62 filaments | | Descriptor: | SEQUESTOSOME-1 | | Authors: | Ciuffa, R, Lamark, T, Tarafder, A, Guesdon, A, Rybina, S, Hagen, W.J.H, Johansen, T, Sachse, C. | | Deposit date: | 2015-03-15 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | The Selective Autophagy Receptor P62 Forms a Flexible Filamentous Helical Scaffold.
Cell Rep., 11, 2015
|
|
4UJD
 
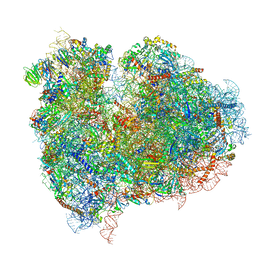 | | mammalian 80S HCV-IRES initiation complex with eIF5B PRE-like state | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Eif5B on Hcv Ires
Nat.Struct.Mol.Biol., 21, 2014
|
|
4UBU
 
 | |
4UHW
 
 | | Human aldehyde oxidase | | Descriptor: | ALDEHYDE OXIDASE, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Coelho, C, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2015-03-26 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Xenobiotic and Inhibitor Binding to Human Aldehyde Oxidase
Nat.Chem.Biol., 11, 2015
|
|
4UCY
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, SULFATE ION, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4UMN
 
 | | Structure of a stapled peptide antagonist bound to Nutlin-resistant Mdm2. | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, M06 | | Authors: | Chee, S, Wongsantichon, J, Quah, S, Robinson, R.C, Verma, C, Lane, D.P, Brown, C.J, Ghadessy, F.J. | | Deposit date: | 2014-05-20 | | Release date: | 2014-05-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of a stapled peptide antagonist bound to nutlin-resistant Mdm2.
PLoS ONE, 9, 2014
|
|
4UOT
 
 | | Thermodynamic hyperstability in parametrically designed helical bundles | | Descriptor: | DESIGNED HELICAL BUNDLE 5H2L | | Authors: | Oberdorfer, G, Huang, P, Pei, X.Y, Xu, C, Gonen, T, Nannenga, B, DiMaio, D, Rogers, J, Luisi, B.F, Baker, D. | | Deposit date: | 2014-06-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High Thermodynamic Stability of Parametrically Designed Helical Bundles
Science, 346, 2014
|
|
9FH9
 
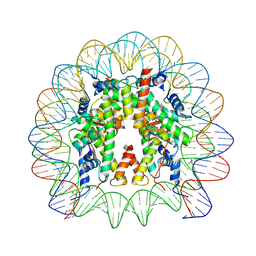 | | Structure of CyclinB1 N-terminus bound to the NCP | | Descriptor: | DNA (145-MER), G2/mitotic-specific cyclin-B1, Histone H2A type 3, ... | | Authors: | Young, R.V.C, Muhammad, R, Alfieri, C. | | Deposit date: | 2024-05-27 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Spatial control of the APC/C ensures the rapid degradation of cyclin B1.
Embo J., 43, 2024
|
|
4UHZ
 
 | | Crystal structure of the human RGMB-BMP2 complex, crystal form 1 | | Descriptor: | BONE MORPHOGENETIC PROTEIN 2, RGM DOMAIN FAMILY MEMBER B, SULFATE ION | | Authors: | Healey, E.G, Bishop, B, Elegheert, J, Bell, C.H, Padilla-Parra, S, Siebold, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Repulsive Guidance Molecule is a Structural Bridge between Neogenin and Bone Morphogenetic Protein.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UIU
 
 | | BROMODOMAIN OF HUMAN BRD9 WITH 7-(3,4-dimethoxyphenyl)-N-(1,1-dioxo-1- thian-4-yl)-5-methyl-4-oxo-4H,5H-thieno-3,2-c-pyridine-2-carboxamide | | Descriptor: | BROMODOMAIN-CONTAINING PROTEIN 9, N-[1,1-bis(oxidanylidene)thian-4-yl]-7-(3,4-dimethoxyphenyl)-5-methyl-4-oxidanylidene-thieno[3,2-c]pyridine-2-carboxamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
9FGQ
 
 | | Structure of human APC3loop 375-381 bound to the NCP | | Descriptor: | Cell division cycle protein 27 homolog, DNA (131-MER), DNA (132-MER), ... | | Authors: | Young, R.V.C, Muhammad, R, Alfieri, C. | | Deposit date: | 2024-05-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Spatial control of the APC/C ensures the rapid degradation of cyclin B1.
Embo J., 43, 2024
|
|
4UBW
 
 | |
4UN3
 
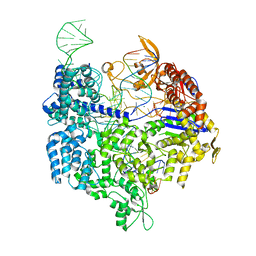 | | Crystal structure of Cas9 bound to PAM-containing DNA target | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
4UCX
 
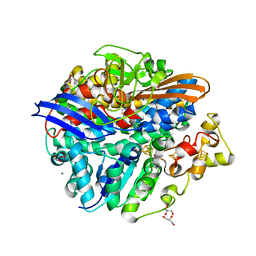 | | Structure of the T18G small subunit mutant of D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Abou-Hamdan, A, Ceccaldi, P, Lebrette, H, Guttierez-Sanz, O, Richaud, P, Cournac, L, Guigliarelli, B, deLacey, A.L, Leger, C, Volbeda, A, Burlat, B, Dementin, S. | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A threonine stabilizes the NiC and NiR catalytic intermediates of [NiFe]-hydrogenase.
J. Biol. Chem., 290, 2015
|
|
4UDV
 
 | | Cryo-EM structure of TMV at 3.35 A resolution | | Descriptor: | 5'-D(*GP*AP*AP)-3', CAPSID PROTEIN | | Authors: | Fromm, S.A, Bharat, T.A.M, Jakobi, A.J, Hagen, W.J.H, Sachse, C. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Seeing Tobacco Mosaic Virus Through Direct Electron Detectors.
J.Struct.Biol., 189, 2015
|
|
4UI2
 
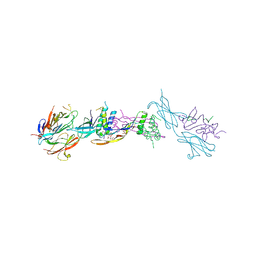 | | Crystal structure of the ternary RGMB-BMP2-NEO1 complex | | Descriptor: | ACETATE ION, BONE MORPHOGENETIC PROTEIN 2, BMP2, ... | | Authors: | Healey, E.G, Bishop, B, Elegheert, J, Bell, C.H, Padilla-Parra, S, Siebold, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Repulsive Guidance Molecule is a Structural Bridge between Neogenin and Bone Morphogenetic Protein.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UIY
 
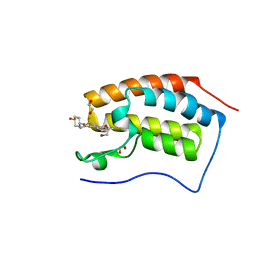 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH N-(1,1-dioxo-1-thian-4-yl)- 5-methyl-4-oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c- pyridine-2-carboximidamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 4, N-[1,1-bis(oxidanylidene)thian-4-yl]-5-methyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
4UCT
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-amino-6-methyl-5-(propan-2-yloxy)-3H-[1,2,4]triazolo[1,5-a]pyrimidin-8-ium, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
7AHK
 
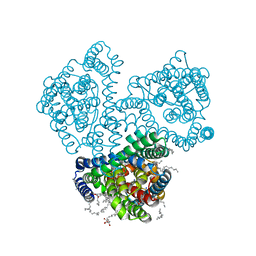 | | Crystal structure of the outward-facing state of the substrate-free Na+-only bound glutamate transporter homolog GltPh | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Alleva, C, Astashkin, A, Machtens, J.-P, Fahlke, C, Gordeliy, V. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Na + -dependent gate dynamics and electrostatic attraction ensure substrate coupling in glutamate transporters.
Sci Adv, 6, 2020
|
|
