3H70
 
 | | Crystal structure of o-succinylbenzoic acid synthetase from staphylococcus aureus Complexed with mg in the active site | | Descriptor: | MAGNESIUM ION, O-succinylbenzoic acid (OSB) synthetase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6QP3
 
 | | Crystal structure of the PLP-bound C-S lyase from Bacillus subtilis (strain 168) | | Descriptor: | ACETATE ION, Cystathionine beta-lyase PatB, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Herman, R, Rudden, M, Wilkinson, A.J, Hanai, S, Thomas, G.H. | | Deposit date: | 2019-02-13 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular basis of thioalcohol production in human body odour.
Sci Rep, 10, 2020
|
|
8JEY
 
 | |
8CTG
 
 | | Extracellular architecture of an engineered canonical Wnt signaling ternary complex | | Descriptor: | Frizzled-8, Low-density lipoprotein receptor-related protein 6, PALMITOLEIC ACID, ... | | Authors: | Tsutsumi, N, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5WWU
 
 | | Crystal Structure of HLA-A*2402 in complex with 2009 pandemic influenza A(H1N1) virus and avian influenza A(H5N1) virus-derived peptide H1-25 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-05 | | Release date: | 2018-01-17 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
5WXD
 
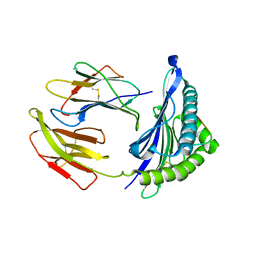 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1) | | Descriptor: | Beta-2-microglobulin, H7-25, HLA class I histocompatibility antigen, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
5W77
 
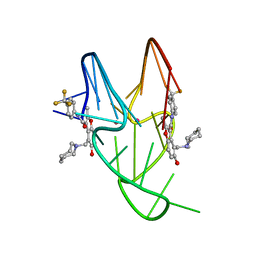 | | Solution structure of the MYC G-quadruplex bound to small molecule DC-34 | | Descriptor: | 4-[(azepan-1-yl)methyl]-5-hydroxy-2-methyl-N-[4-(trifluoromethyl)phenyl]-1-benzofuran-3-carboxamide, DNA (5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3'), POTASSIUM ION | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2017-06-19 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex.
Nat Commun, 9, 2018
|
|
8DWH
 
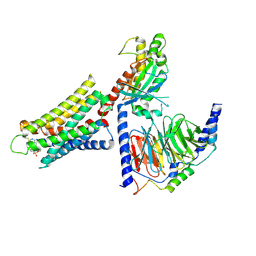 | | CryoEM structure of Gq-coupled MRGPRX1 with ligand Compound-16 | | Descriptor: | Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-01 | | Release date: | 2022-11-02 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
4X20
 
 | | Discovery of cytotoxic Dolastatin 10 analogs with N-terminal modifications | | Descriptor: | 2-methyl-L-prolyl-N-[(3R,4S,5S)-1-{(2S)-2-[(1R,2R)-3-{[(1S)-1-carboxy-2-phenylethyl]amino}-1-methoxy-2-methyl-3-oxopropyl]pyrrolidin-1-yl}-3-methoxy-5-methyl-1-oxoheptan-4-yl]-N-methyl-L-valinamide, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Parris, K.D. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of cytotoxic dolastatin 10 analogues with N-terminal modifications.
J.Med.Chem., 57, 2014
|
|
8DWG
 
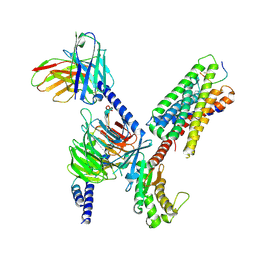 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide ligand BAM8-22 and positive allosteric modulator ML382 | | Descriptor: | 2-[(cyclopropanesulfonyl)amino]-N-(2-ethoxyphenyl)benzamide, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-01 | | Release date: | 2022-11-02 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
8DWC
 
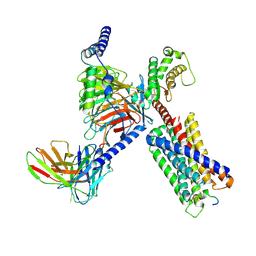 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide agonist BAM8-22 | | Descriptor: | Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-01 | | Release date: | 2022-11-02 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
4X1I
 
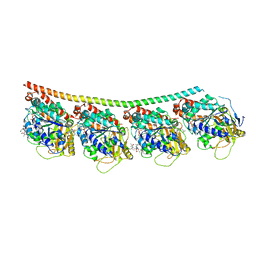 | | Discovery of cytotoxic Dolastatin 10 analogs with N-terminal modifications | | Descriptor: | 2-methyl-L-alanyl-N-[(3R,4S,5S)-3-methoxy-1-{(2S)-2-[(1R,2R)-1-methoxy-2-methyl-3-oxo-3-{[(1S)-2-phenyl-1-(1,3-thiazol-2-yl)ethyl]amino}propyl]pyrrolidin-1-yl}-5-methyl-1-oxoheptan-4-yl]-N-methyl-L-valinamide, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Parris, K.D. | | Deposit date: | 2014-11-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Discovery of cytotoxic dolastatin 10 analogues with N-terminal modifications.
J.Med.Chem., 57, 2014
|
|
3CT2
 
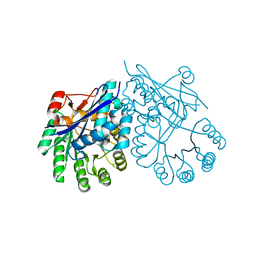 | | Crystal structure of muconate cycloisomerase from Pseudomonas fluorescens | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes.
Biochemistry, 48, 2009
|
|
4X1Y
 
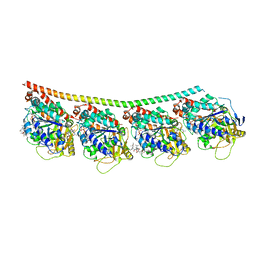 | |
6U7N
 
 | | Crystal structure of neurotrimin (NTM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Neurotrimin, ... | | Authors: | Machius, M, Venkannagari, H, Misra, A, Rudenko, G, Rush, S. | | Deposit date: | 2019-09-03 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.321 Å) | | Cite: | Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes.
J.Mol.Biol., 432, 2020
|
|
6XCR
 
 | | NMR structure of Ost4 in DPC micelles | | Descriptor: | Oligosaccharyltransferase | | Authors: | Chaudhary, B.P. | | Deposit date: | 2020-06-09 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR and MD simulations reveal the impact of the V23D mutation on the function of yeast oligosaccharyltransferase subunit Ost4.
Glycobiology, 31, 2021
|
|
8KHD
 
 | | The interface structure of Omicron RBD binding to 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817, Light chain of 5817, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8KHC
 
 | | SARS-CoV-2 Omicron spike in complex with 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8K0S
 
 | |
3JTY
 
 | | Crystal structure of a BenF-like porin from Pseudomonas fluorescens Pf-5 | | Descriptor: | BenF-like porin, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Sampathkumar, P, Lu, F, Zhao, X, Wasserman, S, Iuzuka, M, Bain, K, Rutter, M, Gheyi, T, Atwell, S, Luz, J, Gilmore, J, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of a putative BenF-like porin from Pseudomonas fluorescens Pf-5 at 2.6 A resolution.
Proteins, 78, 2010
|
|
8K15
 
 | |
8K0R
 
 | |
8K0Q
 
 | |
8K0P
 
 | |
