3FHB
 
 | | Human poly(ADP-ribose) polymerase 3, catalytic fragment in complex with an inhibitor 3-aminobenzoic acid | | Descriptor: | 3-AMINOBENZOIC ACID, Poly [ADP-ribose] polymerase 3 | | Authors: | Lehtio, L, Karlberg, T, Arrowsmith, C.H, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Schueler, H, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3.
J.Med.Chem., 52, 2009
|
|
3N0T
 
 | | Human dipeptidil peptidase DPP7 complexed with inhibitor GSK237826A | | Descriptor: | (3S)-4-oxo-4-piperidin-1-ylbutane-1,3-diamine, Dipeptidyl peptidase 2 | | Authors: | Dobrovetsky, E, Khutoreskaya, G, Seitova, A, Crombet, L, Cossar, D, Pagannon, S, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hassell, A, Shewchuk, L, Haffner, C, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-14 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Human dipeptidyl peptidase DPP7
To be Published
|
|
3II7
 
 | | Crystal structure of the kelch domain of human KLHL7 | | Descriptor: | 1,2-ETHANEDIOL, Kelch-like protein 7 | | Authors: | Chaikuad, A, Thangaratnarajah, C, Cooper, C.D.O, Ugochukwu, E, Muniz, J.R.C, Krojer, T, Sethi, R, Pike, A.C.W, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases.
J.Biol.Chem., 288, 2013
|
|
3H91
 
 | | Crystal structure of the complex of human chromobox homolog 2 (CBX2) and H3K27 peptide | | Descriptor: | Chromobox protein homolog 2, H3K27 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-08-18 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2Z7A
 
 | | X-ray crystal structure of RV0760c from Mycobacterium tuberculosis at 2.10 Angstrom resolution | | Descriptor: | ACETATE ION, Putative steroid isomerase | | Authors: | Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase.
Biochim.Biophys.Acta, 1784, 2008
|
|
3I3C
 
 | | Crystal Structural of CBX5 Chromo Shadow Domain | | Descriptor: | Chromobox protein homolog 5, SODIUM ION | | Authors: | Amaya, M.F, Li, Z, Li, Y, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structural of CBX5 Chromo Shadow Domain
To be Published
|
|
1ZIV
 
 | | Catalytic Domain of Human Calpain-9 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, Calpain 9 | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Mackenzie, F, Dong, A, Choe, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The crystal structures of human calpains 1 and 9 imply diverse mechanisms of action and auto-inhibition.
J.Mol.Biol., 366, 2007
|
|
3NIZ
 
 | | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with ADP bound. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rhodanese family protein | | Authors: | Wernimont, A.K, Dong, A, Lew, J, Lin, Y.H, Hassanali, A, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with ADP bound.
To be Published
|
|
5UBB
 
 | | Crystal structure of human alpha N-terminal protein methyltransferase 1B | | Descriptor: | Alpha N-terminal protein methyltransferase 1B, S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Zhu, L, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An asparagine/glycine switch governs product specificity of human N-terminal methyltransferase NTMT2.
Commun Biol, 1, 2018
|
|
2ARY
 
 | | Catalytic domain of Human Calpain-1 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, Calpain-1 catalytic subunit | | Authors: | Walker, J.R, Davis, T, Lunin, V, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-22 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structures of Human Calpains 1 and 9 Imply Diverse Mechanisms of Action and Auto-inhibition
J.Mol.Biol., 366, 2007
|
|
3IUY
 
 | | Crystal structure of DDX53 DEAD-box domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX53 | | Authors: | Schutz, P, Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
2A7L
 
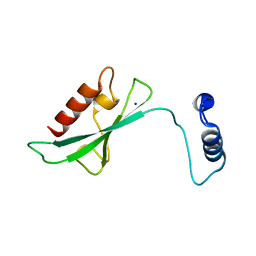 | | Structure of the human hypothetical ubiquitin-conjugating enzyme, LOC55284 | | Descriptor: | Hypothetical ubiquitin-conjugating enzyme LOC55284, SODIUM ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-05 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
3K24
 
 | | Crystal structure of mature apo-Cathepsin L C25A mutant in complex with Gln-Leu-Ala peptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-6)-beta-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin L1, ... | | Authors: | Adams-Cioaba, M.A, Krupa, J.C, Mort, J.S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition and cleavage of histone H3 by cathepsin L.
Nat Commun, 2, 2011
|
|
3FDR
 
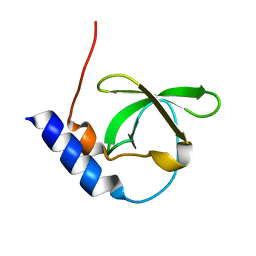 | | Crystal structure of TDRD2 | | Descriptor: | Tudor and KH domain-containing protein | | Authors: | Amaya, M.F, Adams, M.A, Guo, Y, Li, Y, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FL2
 
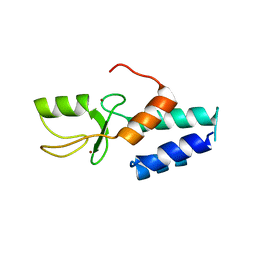 | | Crystal structure of the ring domain of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ring Domain of the E3 Ubiquitin-Protein Ligase Uhrf1
To be Published
|
|
3FM0
 
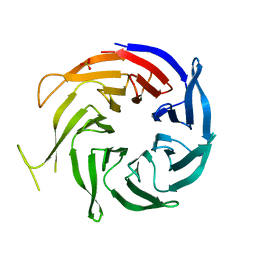 | | Crystal structure of WD40 protein Ciao1 | | Descriptor: | Protein CIAO1, SULFATE ION | | Authors: | Dong, A, Ravichandran, M, Crombet, L, Cossar, D, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
2JM5
 
 | | Solution Structure of the RGS domain from human RGS18 | | Descriptor: | Regulator of G-protein signaling 18 | | Authors: | Higman, V.A, Leidert, M, Bray, J, Elkins, J, Soundararajan, M, Doyle, D.A, Gileadi, C, Phillips, C, Schoch, G, Yang, X, Brockmann, C, Schmieder, P, Diehl, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-11 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3MEA
 
 | | Crystal structure of the SGF29 in complex with H3K4me3 | | Descriptor: | Histone H3, SAGA-associated factor 29 homolog | | Authors: | Bian, C, Xu, C, Tempel, W, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
3K2O
 
 | | Structure of an oxygenase | | Descriptor: | ACETATE ION, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, CHLORIDE ION, ... | | Authors: | Krojer, T, McDonough, M.A, Clifton, I.J, Mantri, M, Ng, S.S, Pike, A.C.W, Butler, D.S, Webby, C.J, Kochan, G, Bhatia, C, Bray, J.E, Chaikuad, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Schofield, C.J, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the 2-Oxoglutarate- and Fe(II)-Dependent Lysyl Hydroxylase JMJD6.
J.Mol.Biol., 401, 2010
|
|
3H9E
 
 | | Crystal structure of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase (GAPDS) complex with NAD and phosphate | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W, Cocking, R, Bray, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure and kinetic characterization of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase, GAPDS.
Biochem.J., 435, 2011
|
|
3GZD
 
 | | Human selenocysteine lyase, P1 crystal form | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, NITRATE ION, Selenocysteine lyase | | Authors: | Karlberg, T, Hogbom, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
2JNU
 
 | | Solution structure of the RGS domain of human RGS14 | | Descriptor: | Regulator of G-protein signaling 14 | | Authors: | Dowler, E.F, Diehl, A, Bray, J, Elkins, J, Soundararajan, M, Doyle, D.A, Gileadi, C, Phillips, C, Schoch, G.A, Yang, X, Brockmann, C, Leidert, M, Rehbein, K, Schmieder, P, Kuhne, R, Higman, V.A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-02 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1GR0
 
 | | myo-inositol 1-phosphate synthase from Mycobacterium tuberculosis in complex with NAD and zinc. | | Descriptor: | CACODYLATE ION, INOSITOL-3-PHOSPHATE SYNTHASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Norman, R.A, Murray-Rust, J, McDonald, N.Q, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-12-10 | | Release date: | 2002-03-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Inositol 1-Phosphate Synthase from Mycobacterium Tuberculosis, a Key Enzyme in Phosphatidylinositol Synthesis
Structure, 10, 2002
|
|
3ME3
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-{[4-(2,3-dihydro-1,4-benzodioxin-6-ylsulfonyl)-1,4-diazepan-1-yl]sulfonyl}aniline, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
3MET
 
 | | Crystal structure of SGF29 in complex with H3K4me2 | | Descriptor: | GLYCEROL, Histone H3, SAGA-associated factor 29 homolog, ... | | Authors: | Bian, C.B, Xu, C, Lam, R, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
