3RZ3
 
 | | Human Cdc34 E2 in complex with CC0651 inhibitor | | Descriptor: | 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, Webb, D.R, Sicheri, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An allosteric inhibitor of the human cdc34 ubiquitin conjugating enzyme
Cell(Cambridge,Mass.), 145, 2011
|
|
5GT2
 
 | | Crystal Structure and Biochemical Features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Probable deferrochelatase/peroxidase YfeX | | Authors: | Ma, Y.L, Yuan, Z.G, Liu, S, Wang, J.X, Gu, L.C, Liu, X.H. | | Deposit date: | 2016-08-18 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Crystal structure and biochemical features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 Asp(143) and Arg(232) play divergent roles toward different substrates
Biochem. Biophys. Res. Commun., 484, 2017
|
|
6M05
 
 | | Trimolecular G-quadruplex | | Descriptor: | DNA (5'-D(*GP*TP*TP*AP*GP*G)-3') | | Authors: | Jing, H.T, Fu, W.Q, Zhang, N. | | Deposit date: | 2020-02-20 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural study on the self-trimerization of d(GTTAGG) into a dynamic trimolecular G-quadruplex assembly preferentially in Na+ solution with a moderate K+ tolerance.
Nucleic Acids Res., 49, 2021
|
|
8F22
 
 | |
7Y4I
 
 | | Crystal structure of SPINDLY in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Xu, S.T, Wan, L.H. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into mechanism and specificity of the plant protein O-fucosyltransferase SPINDLY.
Nat Commun, 13, 2022
|
|
7R7V
 
 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 | | Descriptor: | Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, GLYCEROL, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7Y
 
 | | Crystal structure of HLA-B*5701 complex with an HIV-1 Gag-derived epitope QW9 S3T variant | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, GLYCEROL, ... | | Authors: | Li, X.L, Ng, R, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7W
 
 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 S3T variant | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, MHC class I antigen | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7X
 
 | | Crystal structure of HLA-B*5701 complex with an HIV-1 Gag-derived epitope QW9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R80
 
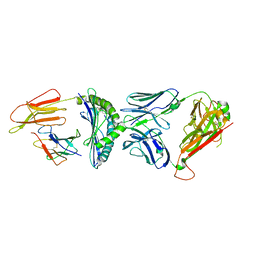 | | Crystal structure of C3 TCR complex with QW9-bound HLA-B*5301 | | Descriptor: | Alpha chain of C3 TCR, Beta Chain of C3 TCR, Beta-2-microglobulin, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7SH7
 
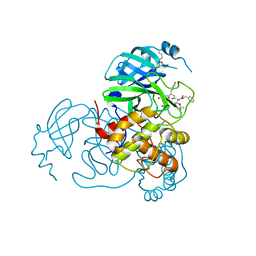 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI87 | | Descriptor: | 3C-like proteinase nsp5, benzyl [(2S,3R)-3-tert-butoxy-1-{[(2S)-3-cyclohexyl-1-oxo-1-(2-{[(3S)-2-oxopyrrolidin-3-yl]methyl}-2-propanoylhydrazinyl)propan-2-yl]amino}-1-oxobutan-2-yl]carbamate (non-preferred name) | | Authors: | Blankenship, L.R, Yang, K.S, Liu, W.R. | | Deposit date: | 2021-10-08 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Azapeptide Platform in Conjunction with Covalent Warheads to Uncover High-Potency Inhibitors for SARS-CoV-2 Main Protease.
Biorxiv, 2023
|
|
6CTE
 
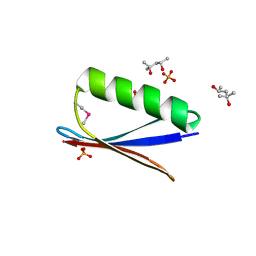 | | 77Se-NMR probes the protein environment of selenomethionine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Chen, Q, Rozovsky, S. | | Deposit date: | 2018-03-22 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
6CPZ
 
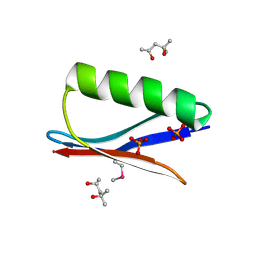 | | Selenomethionine mutant (I6Sem) of protein GB1 examined by X-ray diffraction | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin G-binding protein G, ... | | Authors: | Chen, Q, Rozovsky, S. | | Deposit date: | 2018-03-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
7SDA
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI49 | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4,4-dimethyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
7SD9
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI48 | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4,4-dimethyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
7SDC
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MI-09 | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-{[4-(trifluoromethoxy)phenoxy]acetyl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
5HXB
 
 | | Cereblon in complex with DDB1, CC-885, and GSPT1 | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)urea, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | Authors: | Chamberlain, P.P, Matyskiela, M, Pagarigan, B. | | Deposit date: | 2016-01-30 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase.
Nature, 535, 2016
|
|
4QNC
 
 | | Crystal structure of a SemiSWEET in an occluded state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PENTADECANE, chemical transport protein | | Authors: | Yan, X, Yuyong, T, Liang, F, Perry, K. | | Deposit date: | 2014-06-17 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structures of bacterial homologues of SWEET transporters in two distinct conformations.
Nature, 515, 2014
|
|
3GFE
 
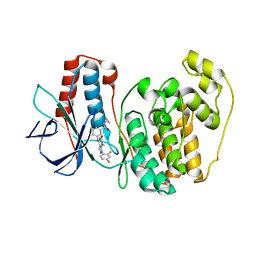 | | Crystal Structure of p38a Mitogen-Activated Protein Kinase in Complex with a Pyrazolopyridinone Inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-3-{[1-(2,4-difluorophenyl)-7-methyl-6-oxo-6,7-dihydro-1H-pyrazolo[3,4-b]pyridin-4-yl]amino}-4-methylbenzamide | | Authors: | Mohr, C, Jordan, S. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Part 1: Structure-Activity Relationship (SAR) investigations of fused pyrazoles as potent, selective and orally available inhibitors of p38alpha mitogen-activated protein kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6C9O
 
 | |
6CHE
 
 | |
8ACL
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GC-14 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-pyridin-3-ylcarbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
6CNE
 
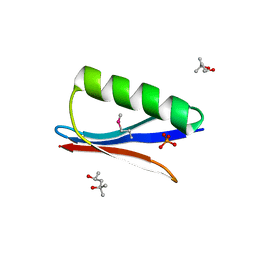 | | Selenomethionine variant (V29SeM) of protein GB1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin G-binding protein G, PHOSPHATE ION | | Authors: | Chen, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
8ACD
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GA-17S | | Descriptor: | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
6OC0
 
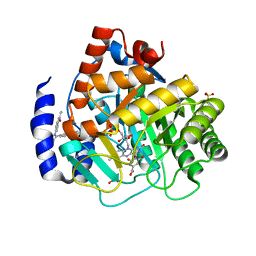 | | Crystal structure of human DHODH with OSU-03012 | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Durst, M.A, Lavie, A. | | Deposit date: | 2019-03-21 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Metabolic Modifier Screen Reveals Secondary Targets of Protein Kinase Inhibitors within Nucleotide Metabolism.
Cell Chem Biol, 27, 2020
|
|
