7Y62
 
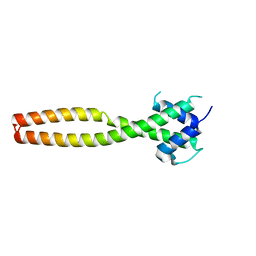 | | Crystal structure of human TFEB HLHLZ domain | | Descriptor: | Transcription factor EB | | Authors: | Yang, G, Li, P, Lin, Y, Liu, Z, Sun, H, Zhao, Z, Fang, P, Wang, J. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A small-molecule drug inhibits autophagy gene expression through the central regulator TFEB.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7A96
 
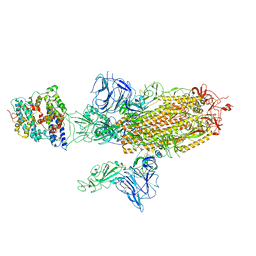 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Anticlockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A97
 
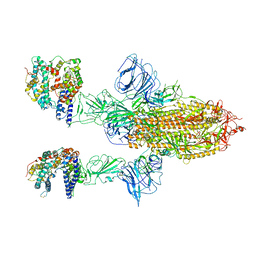 | | SARS-CoV-2 Spike Glycoprotein with 2 ACE2 Bound | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A93
 
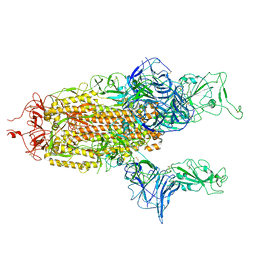 | |
7A98
 
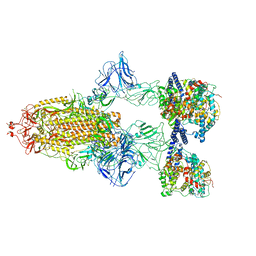 | | SARS-CoV-2 Spike Glycoprotein with 3 ACE2 Bound | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7BNN
 
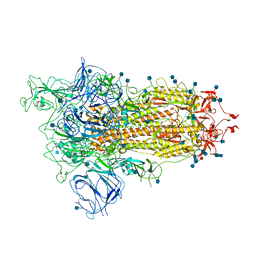 | | Open conformation of D614G SARS-CoV-2 spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BBH
 
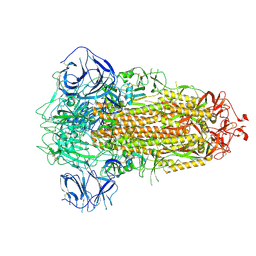 | | Structure of Coronavirus Spike from Smuggled Guangdong Pangolin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface glycoprotein | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7A92
 
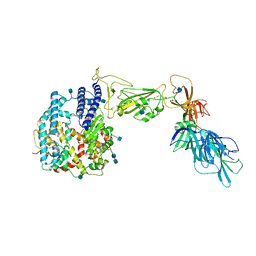 | | Dissociated S1 domain of SARS-CoV-2 Spike bound to ACE2 (Unmasked Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-30 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A94
 
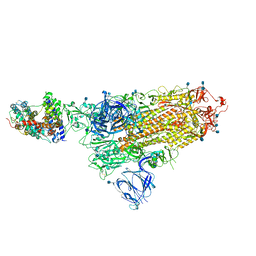 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A91
 
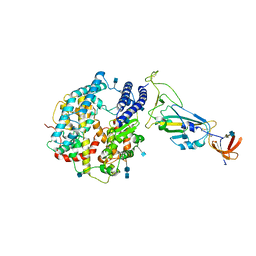 | | Dissociated S1 domain of SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A95
 
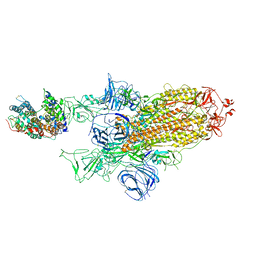 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Clockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7BNM
 
 | | Closed conformation of D614G SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BNO
 
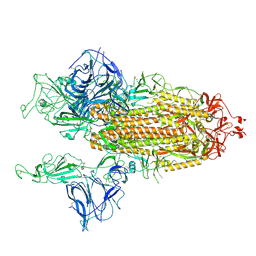 | | Open conformation of D614G SARS-CoV-2 spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4UEI
 
 | | Solution structure of the sterol carrier protein domain 2 of Helicoverpa armigera | | Descriptor: | STEROL CARRIER PROTEIN 2/3-OXOACYL-COA THIOLASE | | Authors: | Liu, X, Ma, H, Yan, X, Hong, H, Peng, J, Peng, R. | | Deposit date: | 2014-12-18 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Function of Helicoverpa Armigera Sterol Carrier Protein-2, an Important Insecticidal Target from the Cotton Bollworm.
Sci.Rep., 5, 2015
|
|
3LRV
 
 | |
3NMW
 
 | | Crystal structure of armadillo repeats domain of APC | | Descriptor: | APC variant protein, SULFATE ION | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-22 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
4DEQ
 
 | | Structure of the Neuropilin-1/VEGF-A complex | | Descriptor: | Neuropilin-1, Vascular endothelial growth factor A, PHOSPHATE ION | | Authors: | Vander Kooi, C.W. | | Deposit date: | 2012-01-21 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Structural Basis for Selective Vascular Endothelial Growth Factor-A (VEGF-A) Binding to Neuropilin-1.
J.Biol.Chem., 287, 2012
|
|
5WXP
 
 | | Crystal structure of uPA in complex with upain-2-3-W3A | | Descriptor: | ALANINE, CYSTEINE, Urokinase-type plasminogen activator chain B, ... | | Authors: | Jiang, L, Huang, M. | | Deposit date: | 2017-01-08 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cleavage of peptidic inhibitors by target protease is caused by peptide conformational transition.
Biochim. Biophys. Acta, 1862, 2018
|
|
3RL7
 
 | | Crystal structure of hDLG1-PDZ1 complexed with APC | | Descriptor: | 11-mer peptide from Adenomatous polyposis coli protein, Disks large homolog 1 | | Authors: | Zhang, Z, Li, H, Wu, G. | | Deposit date: | 2011-04-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein.
Plos One, 6, 2011
|
|
3RL8
 
 | | Crystal structure of hDLG1-PDZ2 complexed with APC | | Descriptor: | 11-mer peptide from Adenomatous polyposis coli protein, Disks large homolog 1 | | Authors: | Zhang, Z, Li, H, Wu, G. | | Deposit date: | 2011-04-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein.
Plos One, 6, 2011
|
|
5WXF
 
 | | Crystal structure of uPA in complex with upain-2-2 | | Descriptor: | SULFATE ION, Urokinase-type plasminogen activator chain B, upain-2-2 peptide | | Authors: | Jiang, L, Huang, M. | | Deposit date: | 2017-01-07 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Cleavage of peptidic inhibitors by target protease is caused by peptide conformational transition.
Biochim. Biophys. Acta, 1862, 2018
|
|
5WXO
 
 | | Crystal structure of uPA in complex with upain-2-2-W3A | | Descriptor: | Urokinase-type plasminogen activator chain B, upain-2-2-W3A peptide | | Authors: | Jiang, L, Huang, M. | | Deposit date: | 2017-01-08 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Cleavage of peptidic inhibitors by target protease is caused by peptide conformational transition.
Biochim. Biophys. Acta, 1862, 2018
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
