2KAN
 
 | | Solution NMR structure of ubiquitin-like domain of Arabidopsis thaliana protein At2g32350. Northeast Structural Genomics Consortium target AR3433A | | Descriptor: | uncharacterized protein ar3433a | | Authors: | Eletsky, A, Mills, J.L, Sukumaran, D, Hua, J, Shastry, R, Jiang, M, Ciccosanti, C, Xiao, R, Liu, J, Everret, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-11 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of ubiquitin-like domain of Arabidopsis thaliana protein At2g32350.
To be Published
|
|
2K5H
 
 | | Solution NMR structure of protein encoded by MTH693 from Methanobacterium thermoautotrophicum: Northeast Structural Genomics Consortium target tt824a | | Descriptor: | Conserved protein | | Authors: | Wu, Y, Singarapu, K, Semesi, A, Sukumaran, D, Yee, A, Garcia, M, Arrowsmith, C, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein encoded by MTH693 from Methanobacterium thermoautotrophicum: Northeast Structural Genomics Consortium target tt824a
To be Published
|
|
2K4M
 
 | | Solution NMR Structure of M. thermoautotrophicum protein MTH_1000, Northeast Structural Genomics Consortium Target TR8 | | Descriptor: | UPF0146 protein MTH_1000 | | Authors: | Eletsky, A, Sukumaran, D, Semesi, A, Guido, V, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-13 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of M. thermoautotrophicum protein MTH_1000, Northeast Structural Genomics Consortium Target TR8
To be Published
|
|
2KEL
 
 | | Structure of the transcription regulator SvtR from the hyperthermophilic archaeal virus SIRV1 | | Descriptor: | Uncharacterized protein 56B | | Authors: | Guilliere, F, Kessler, A, Peixeiro, N, Sezonov, G, Prangishvili, D, Delepierre, M, Guijarro, J.I. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1.
J.Biol.Chem., 284, 2009
|
|
3BZW
 
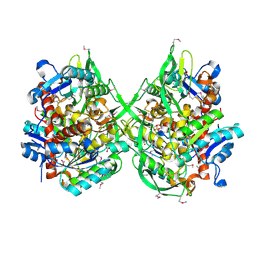 | | Crystal structure of a putative lipase from Bacteroides thetaiotaomicron | | Descriptor: | ACETATE ION, Putative lipase, SULFATE ION | | Authors: | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-18 | | Release date: | 2008-02-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of a putative lipase from Bacteroides thetaiotaomicron.
To be Published
|
|
2QVC
 
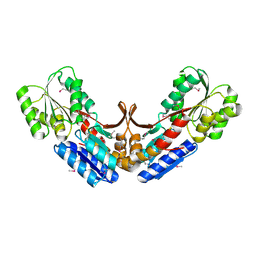 | | Crystal structure of a periplasmic sugar ABC transporter from Thermotoga maritima | | Descriptor: | Sugar ABC transporter, periplasmic sugar-binding protein, beta-D-glucopyranose | | Authors: | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a periplasmic glucose-binding protein from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2W8F
 
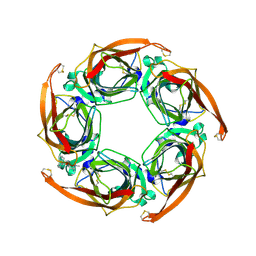 | | Aplysia californica AChBP bound to in silico compound 31 | | Descriptor: | (3-EXO)-3-(10,11-DIHYDRO-5H-DIBENZO[A,D][7]ANNULEN-5-YLOXY)-8,8-DIMETHYL-8-AZONIABICYCLO[3.2.1]OCTANE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Edink, E, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, de Esch, I.J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X- Ray Analysis.
J.Med.Chem., 52, 2009
|
|
4XI0
 
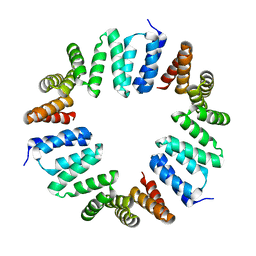 | | MamA 41-end from Desulfovibrio magneticus RS-1 | | Descriptor: | Magnetosome protein MamA | | Authors: | Zarivach, R, Zeytuni, N, Cronin, S, Davidov, G, Baran, D, Stein, T. | | Deposit date: | 2015-01-06 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | MamA as a Model Protein for Structure-Based Insight into the Evolutionary Origins of Magnetotactic Bacteria.
Plos One, 10, 2015
|
|
3HH7
 
 | | Structural and Functional Characterization of a Novel Homodimeric Three-finger Neurotoxin from the Venom of Ophiophagus hannah (King Cobra) | | Descriptor: | Muscarinic toxin-like protein 3 homolog | | Authors: | Roy, A, Zhou, X, Chong, M.Z, D'hoedt, D, Foo, C.S, Rajagopalan, N, Nirthanan, S, Bertrand, D, Sivaraman, J, Kini, R.M. | | Deposit date: | 2009-05-15 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Functional Characterization of a Novel Homodimeric Three-finger Neurotoxin from the Venom of Ophiophagus hannah (King Cobra)
J.Biol.Chem., 285, 2010
|
|
3P27
 
 | | Crystal structure of S. cerevisiae Hbs1 protein (GDP-bound form), a translational GTPase involved in RNA quality control pathways and interacting with Dom34/Pelota | | Descriptor: | Elongation factor 1 alpha-like protein, GUANOSINE-5'-DIPHOSPHATE | | Authors: | van den Elzen, A, Henri, J, Lazar, N, Gas, M.E, Durand, D, Lacroute, F, Nicaise, M, van Tilbeurgh, H, Sraphin, B, Graille, M, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2010-10-01 | | Release date: | 2010-11-17 | | Last modified: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3D3A
 
 | |
2LVH
 
 | | Solution structure of the zinc finger AFV1p06 protein from the hyperthermophilic archaeal virus AFV1 | | Descriptor: | Putative zinc finger protein ORF59a, ZINC ION | | Authors: | Guilliere, F, Sezonov, G, Prangishvili, D, Delepierre, M, Guijarro, J. | | Deposit date: | 2012-07-05 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an archaeal DNA binding protein with an eukaryotic zinc finger fold.
Plos One, 8, 2013
|
|
4YDU
 
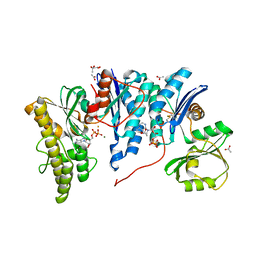 | | Crystal structure of E. coli YgjD-YeaZ heterodimer in complex with ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Zhang, W, Collinet, B, Perrochia, L, Durand, D, Van Tilbeurgh, H. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
2QXY
 
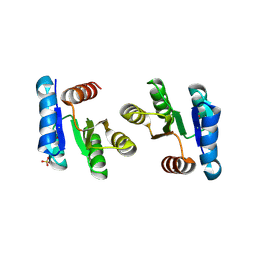 | |
2J9N
 
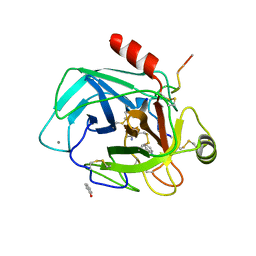 | | Robotically harvested Trypsin complexed with Benzamidine containing polypeptide mediated crystal contacts | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Viola, R, Carman, P, Walsh, J, Miller, E, Benning, M, Frankel, D, McPherson, A, Cudney, R, Rupp, B. | | Deposit date: | 2006-11-14 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Operator Assisted Harvesting of Protein Crystals Using a Universal Micromanipulation Robot.
J.Appl.Crystallogr., 40, 2007
|
|
2MC1
 
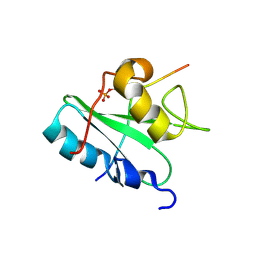 | | Solution structure of the Vav1 SH2 domain complexed with a Syk-derived singly phosphorylated peptide | | Descriptor: | Proto-oncogene vav, Tyrosine-protein kinase SYK | | Authors: | Chen, C, Piraner, D, Gorenstein, N.M, Geahlen, R.L, Post, C.B. | | Deposit date: | 2013-08-13 | | Release date: | 2013-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Differential recognition of syk-binding sites by each of the two phosphotyrosine-binding pockets of the Vav SH2 domain.
Biopolymers, 99, 2013
|
|
2SPC
 
 | | CRYSTAL STRUCTURE OF THE REPETITIVE SEGMENTS OF SPECTRIN | | Descriptor: | SPECTRIN | | Authors: | Yan, Y, Winograd, E, Viel, A, Cronin, T, Harrison, S.C, Branton, D. | | Deposit date: | 1994-03-01 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the repetitive segments of spectrin.
Science, 262, 1993
|
|
3DEB
 
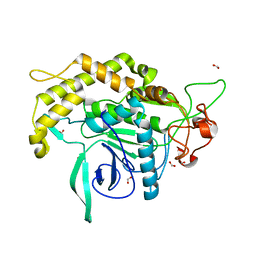 | |
6NAV
 
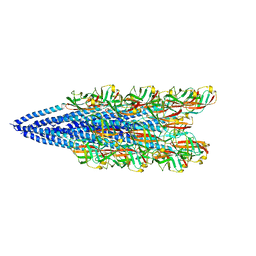 | | Cryo-EM reconstruction of Sulfolobus islandicus LAL14/1 Pilus | | Descriptor: | M9UD72 | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2018-12-06 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | An extensively glycosylated archaeal pilus survives extreme conditions.
Nat Microbiol, 4, 2019
|
|
2JV8
 
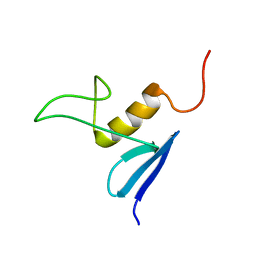 | | Solution structure of protein NE1242 from Nitrosomonas europaea. Northeast Structural Genomics Consortium Target NeT4 | | Descriptor: | Uncharacterized protein NE1242 | | Authors: | Wu, Y, Yee, A, Zeri, A.C, Guido, V, Sukumaran, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-12 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein NE1242 from Nitrosomonas europaea.
To be Published
|
|
3OVG
 
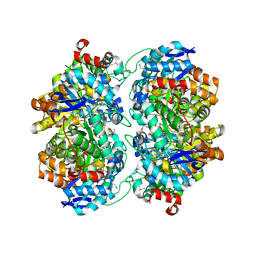 | | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound | | Descriptor: | PHOSPHATE ION, ZINC ION, amidohydrolase | | Authors: | Zhang, Z, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound
To be Published
|
|
2VDV
 
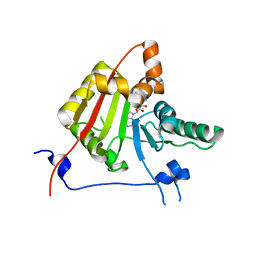 | | Structure of trm8, m7G methylation enzyme | | Descriptor: | S-ADENOSYLMETHIONINE, TRNA (GUANINE-N(7)-)-METHYLTRANSFERASE | | Authors: | Leulliot, N, Chaillet, M, Durand, D, Ulryck, N, Blondeau, K, Van Tilbeurgh, H. | | Deposit date: | 2007-10-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Yeast tRNA M7G Methylation Complex.
Structure, 16, 2008
|
|
6TAZ
 
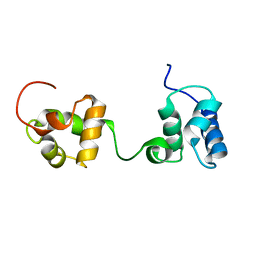 | | Timeless couples G quadruplex detection with processing by DDX11 during DNA replication | | Descriptor: | Protein timeless homolog | | Authors: | Lerner Koch, L, Holzer, S, Kilkenny, M.L, Murat, P, Svikovic, S, Schiavone, D, Bittleston, A, Maman, J.D, Branzei, D, Stott, K, Pellegrini, L, Sale, E.J. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Timeless couples G-quadruplex detection with processing by DDX11 helicase during DNA replication.
Embo J., 39, 2020
|
|
6UAN
 
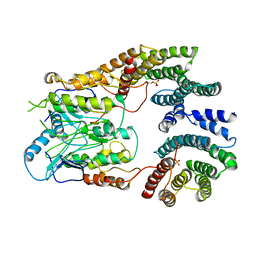 | | B-Raf:14-3-3 complex | | Descriptor: | 14-3-3 zeta, Serine/threonine-protein kinase B-raf | | Authors: | Kondo, Y, Ognjenovic, J, Banerjee, S, Karandur, D, Merk, A, Kulhanek, K, Wong, K, Roose, J.P, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Science, 366, 2019
|
|
2JZ6
 
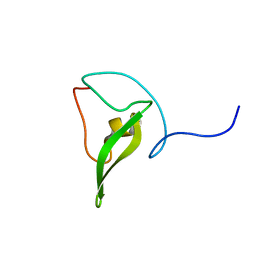 | | Solution structure of 50S ribosomal protein L28 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR97 | | Descriptor: | 50S ribosomal protein L28 | | Authors: | Wu, Y, Singarapu, K.K, Guido, V, Yee, A, Sukumaran, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-01-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 50S ribosomal protein L28 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR97.
To be Published
|
|
