1KMZ
 
 | | MOLECULAR BASIS OF MITOMYCIN C RESICTANCE IN STREPTOMYCES: CRYSTAL STRUCTURES OF THE MRD PROTEIN WITH AND WITHOUT A DRUG DERIVATIVE | | Descriptor: | mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1KLL
 
 | | Molecular basis of mitomycin C resictance in streptomyces: Crystal structures of the MRD protein with and without a drug derivative | | Descriptor: | 1,2-CIS-1-HYDROXY-2,7-DIAMINO-MITOSENE, mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-12 | | Release date: | 2002-07-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1LQV
 
 | | Crystal structure of the Endothelial protein C receptor with phospholipid in the groove in complex with Gla domain of protein C. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-05-13 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
1C5E
 
 | | BACTERIOPHAGE LAMBDA HEAD PROTEIN D | | Descriptor: | GLYCEROL, HEAD DECORATION PROTEIN | | Authors: | Yang, F, Forrer, P, Dauter, Z, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1999-11-18 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD.
Nat.Struct.Biol., 7, 2000
|
|
1AYX
 
 | | CRYSTAL STRUCTURE OF GLUCOAMYLASE FROM SACCHAROMYCOPSIS FIBULIGERA AT 1.7 ANGSTROMS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUCOAMYLASE | | Authors: | Sevcik, J, Hostinova, E, Gasperik, J, Solovicova, A, Wilson, K.S, Dauter, Z. | | Deposit date: | 1997-11-12 | | Release date: | 1998-05-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of glucoamylase from Saccharomycopsis fibuligera at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AY7
 
 | | RIBONUCLEASE SA COMPLEX WITH BARSTAR | | Descriptor: | BARSTAR, GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Sevcik, J, Urbanikova, L, Dauter, Z, Wilson, K.S. | | Deposit date: | 1997-11-14 | | Release date: | 1999-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of RNase Sa by the inhibitor barstar: structure of the complex at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1BWW
 
 | | BENCE-JONES IMMUNOGLOBULIN REI VARIABLE PORTION, T39K MUTANT | | Descriptor: | PROTEIN (IG KAPPA CHAIN V-I REGION REI) | | Authors: | Uson, I, Pohl, E, Schneider, T.R, Dauter, Z, Schmidt, A, Fritz, H.J, Sheldrick, G.M. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 A structure of the stabilized REIv mutant T39K. Application of local NCS restraints.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BOX
 
 | | N39S MUTANT OF RNASE SA FROM STREPTOMYCES AUREOFACIENS | | Descriptor: | GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Hebert, E.J, Giletto, A, Sevcik, J, Urbanikova, L, Wilson, K.S, Dauter, Z, Pace, C.N. | | Deposit date: | 1998-08-07 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Contribution of a conserved asparagine to the conformational stability of ribonucleases Sa, Ba, and T1.
Biochemistry, 37, 1998
|
|
1CC0
 
 | | CRYSTAL STRUCTURE OF THE RHOA.GDP-RHOGDI COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, rho GDP dissociation inhibitor alpha, ... | | Authors: | Longenecker, K.L, Read, P, Derewenda, U, Dauter, Z, Garrard, S, Walker, L, Somlyo, A.V, Somlyo, A.P, Nakamoto, R.K, Derewenda, Z.S. | | Deposit date: | 1999-03-03 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | How RhoGDI binds Rho.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1L8J
 
 | | Crystal Structure of the Endothelial Protein C Receptor and Bound Phospholipid Molecule | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-03-20 | | Release date: | 2002-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
1LNI
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS AT ATOMIC RESOLUTION (1.0 A) | | Descriptor: | GLYCEROL, GUANYL-SPECIFIC RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Lamzin, V.S, Dauter, Z, Wilson, K.S. | | Deposit date: | 2002-05-03 | | Release date: | 2002-07-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution data reveal flexibility in the structure of RNase Sa.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1CTN
 
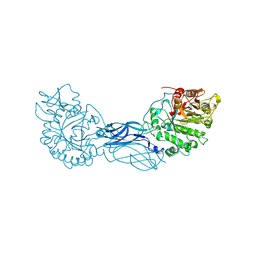 | | CRYSTAL STRUCTURE OF A BACTERIAL CHITINASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CHITINASE A | | Authors: | Perrakis, A, Tews, I, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bacterial chitinase at 2.3 A resolution.
Structure, 2, 1994
|
|
1CXQ
 
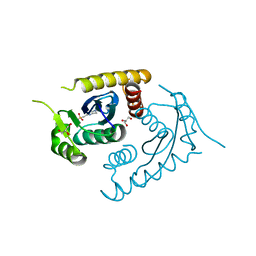 | | ATOMIC RESOLUTION ASV INTEGRASE CORE DOMAIN FROM AMMONIUM SULFATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE, GLYCEROL | | Authors: | Lubkowski, J, Dauter, Z, Yang, F, Alexandratos, J, Merkel, G, Skalka, A.M, Wlodawer, A. | | Deposit date: | 1999-08-30 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant.
Biochemistry, 38, 1999
|
|
1CZB
 
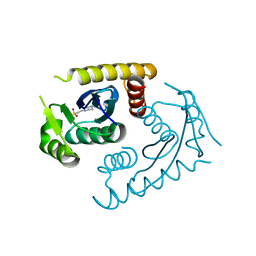 | | ATOMIC RESOLUTION ASV INTEGRASE CORE DOMAIN FROM HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE | | Authors: | Lubkowski, J, Dauter, Z, Yang, F, Alexandratos, J, Merkel, G, Skalka, A.M, Wlodawer, A. | | Deposit date: | 1999-09-01 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant.
Biochemistry, 38, 1999
|
|
1X3E
 
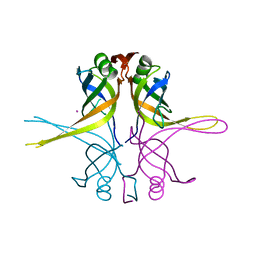 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium smegmatis | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2005-05-04 | | Release date: | 2005-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1X3G
 
 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium SMEGMATIS | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1WQ6
 
 | | The tetramer structure of the nervy homolgy two (NHR2) domain of AML1-ETO is critical for AML1-ETO'S activity | | Descriptor: | AML1-ETO | | Authors: | Liu, Y, Cheney, M.D, Chruszcz, M, Lukasik, S.M, Hartman, K.L, Laue, T.M, Dauter, Z, Minor, W, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2004-09-23 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity
Cancer Cell, 9, 2006
|
|
1X3F
 
 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium SMEGMATIS | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1XCG
 
 | | Crystal Structure of Human RhoA in complex with DH/PH fragment of PDZRHOGEF | | Descriptor: | Rho guanine nucleotide exchange factor 11, Transforming protein RhoA | | Authors: | Derewenda, U, Oleksy, A, Stevenson, A.S, Korczynska, J, Dauter, Z, Somlyo, A.P, Otlewski, J, Somlyo, A.V, Derewenda, Z.S. | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca(2+) sensitization pathway in smooth muscle
Structure, 12, 2004
|
|
1VZY
 
 | | Crystal structure of the Bacillus subtilis HSP33 | | Descriptor: | 33 KDA CHAPERONIN, ACETATE ION, ZINC ION | | Authors: | Janda, I.K, Devedjiev, Y, Derewenda, U, Dauter, Z, Bielnicki, J, Cooper, D.R, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-29 | | Release date: | 2004-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of the reduced, Zn2+-bound form of the B. subtilis Hsp33 chaperone and its implications for the activation mechanism.
Structure, 12, 2004
|
|
1YKQ
 
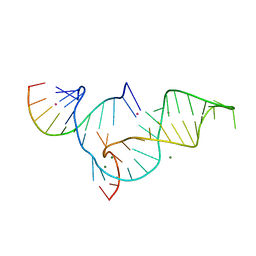 | | Crystal structure of Diels-Alder ribozyme | | Descriptor: | CADMIUM ION, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YKV
 
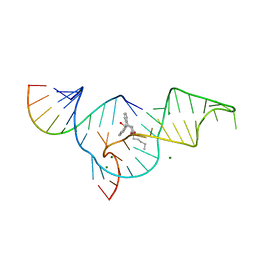 | | Crystal structure of the Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YLS
 
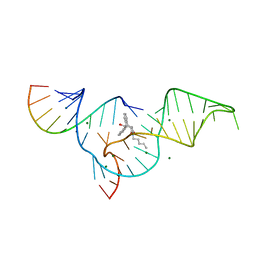 | | Crystal structure of selenium-modified Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, MAGNESIUM ION, RNA Diels-Alder ribozyme | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1WAB
 
 | | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | ACETATE ION, PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Swenson, L, Derewenda, U, Serre, L, Wei, Y, Dauter, Z, Hattori, M, Aoki, J, Arai, H, Adachi, T, Inoue, K, Derewenda, Z.S. | | Deposit date: | 1996-10-30 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Brain acetylhydrolase that inactivates platelet-activating factor is a G-protein-like trimer.
Nature, 385, 1997
|
|
1W9X
 
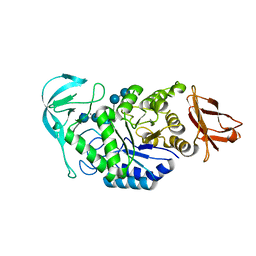 | | Bacillus halmapalus alpha amylase | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA AMYLASE, CALCIUM ION, ... | | Authors: | Davies, G.J, Brzozowski, A.M, Dauter, Z, Rasmussen, M.D, Borchert, T.V, Wilson, K.S. | | Deposit date: | 2004-10-20 | | Release date: | 2005-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Bacillus Halmapalus Family 13 Alpha-Amylase, Bha, in Complex with an Acarbose-Derived Nonasaccharide at 2.1 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
