6N4Y
 
 | | Metabotropic Glutamate Receptor 5 Extracellular Domain with Nb43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Chu, M, Weis, W.I, Mathiesen, J.M, Skiniotis, G, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.262 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
2LNL
 
 | | Structure of human CXCR1 in phospholipid bilayers | | Descriptor: | C-X-C chemokine receptor type 1 | | Authors: | Park, S, Das, B.B, Casagrande, F, Nothnagel, H, Chu, M, Kiefer, H, Maier, K, De Angelis, A, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-12-31 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the chemokine receptor CXCR1 in phospholipid bilayers.
Nature, 491, 2012
|
|
4USO
 
 | | X-ray structure of the CCL2 lectin in complex with sialyl lewis X | | Descriptor: | CCL2 LECTIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
4USP
 
 | | X-ray structure of the dimeric CCL2 lectin in native form | | Descriptor: | CCL2 LECTIN, CHLORIDE ION, PHOSPHATE ION | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
2OVW
 
 | | ENDOGLUCANASE I COMPLEXED WITH CELLOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Sulzenbacher, G, Davies, G.J, Schulein, M. | | Deposit date: | 1997-04-04 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the endoglucanase I from Fusarium oxysporum: native, cellobiose, and 3,4-epoxybutyl beta-D-cellobioside-inhibited forms, at 2.3 A resolution.
Biochemistry, 36, 1997
|
|
2OM7
 
 | | Structural Basis for Interaction of the Ribosome with the Switch Regions of GTP-bound Elongation Factors | | Descriptor: | 16S ribosomal RNA (H5), 30S ribosomal protein S12, 30S ribosomal protein S2, ... | | Authors: | Connell, S.R, Wilson, D.N, Rost, M, Schueler, M, Giesebrecht, J, Dabrowski, M, Mielke, T, Fucini, P, Spahn, C.M.T. | | Deposit date: | 2007-01-21 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors.
Mol.Cell, 25, 2007
|
|
1U36
 
 | | Crystal structure of WLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3Y
 
 | | Crystal structure of ILAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U42
 
 | | Crystal structure of MLAM mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U41
 
 | | Crystal structure of YLGV mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3Z
 
 | | Crystal structure of MLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3J
 
 | | Crystal structure of MLAV mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
6EU9
 
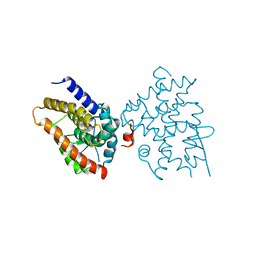 | | Crystal structure of Platynereis dumerilii RAR ligand-binding domain in complex with all-trans retinoic acid | | Descriptor: | RETINOIC ACID, Retinoic acid receptor | | Authors: | Handberg-Thorsager, M, Gutierrez-Mazariegos, J, Arold, S.T, Nadendla, E.K, Bertucci, P.Y, Germain, P, Tomancak, P, Pierzchalski, K, Jones, J.W, Albalat, R, Kane, M.A, Bourguet, W, Laudet, V, Arendt, D, Schubert, M. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The ancestral retinoic acid receptor was a low-affinity sensor triggering neuronal differentiation.
Sci Adv, 4, 2018
|
|
6A3H
 
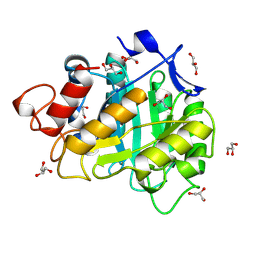 | | 2-DEOXY-2-FLURO-B-D-CELLOTRIOSYL/ENZYME INTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.6 ANGSTROM RESOLUTION | | Descriptor: | ENDOGLUCANASE, GLYCEROL, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-22 | | Release date: | 1999-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
7A3H
 
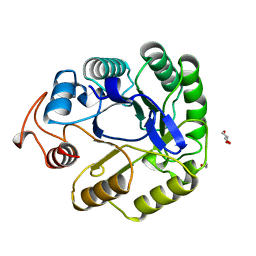 | | NATIVE ENDOGLUCANASE CEL5A CATALYTIC CORE DOMAIN AT 0.95 ANGSTROMS RESOLUTION | | Descriptor: | ENDOGLUCANASE, ETHANOL, GLYCEROL | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-08-05 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
8A0C
 
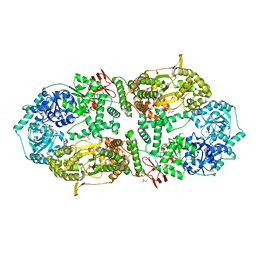 | | Capsular polysaccharide synthesis multienzyme in complex with CMP | | Descriptor: | Bcs3, CYTIDINE-5'-MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Cifuente, J.O, Schulze, J, Bethe, A, Di Domenico, V, Litschko, C, Budde, I, Eidenberger, L, Thiesler, H, Ramon-Roth, I, Berger, M, Claus, H, DAngelo, C, Marina, A, Gerardy-Schahn, R, Schubert, M, Guerin, M.E, Fiebig, T. | | Deposit date: | 2022-05-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A multi-enzyme machine polymerizes the Haemophilus influenzae type b capsule.
Nat.Chem.Biol., 19, 2023
|
|
8A0M
 
 | | Capsular polysaccharide synthesis multienzyme in complex with capsular polymer fragment | | Descriptor: | Bcs3, MAGNESIUM ION, beta-D-ribosyl-(1->1)-D-ribitol-5-phosphate | | Authors: | Cifuente, J.O, Schulze, J, Bethe, A, Di Domenico, V, Litschko, C, Budde, I, Eidenberger, L, Thiesler, H, Ramon-Roth, I, Berger, M, Claus, H, DAngelo, C, Marina, A, Gerardy-Schahn, R, Schubert, M, Guerin, M.E, Fiebig, T. | | Deposit date: | 2022-05-29 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A multi-enzyme machine polymerizes the Haemophilus influenzae type b capsule.
Nat.Chem.Biol., 19, 2023
|
|
6QHE
 
 | | Alcohol Dehydrogenase from Arthrobacter sp. TS-15 in complex with NAD+ | | Descriptor: | Alcohol Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Lockie, C, Beloti, L, Shanati, T, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2019-01-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Two Enantiocomplementary Ephedrine Dehydrogenases from Arthrobacter sp. TS-15 with Broad Substrate Specificity
Acs Catalysis, 9, 2019
|
|
4ENG
 
 | | STRUCTURE OF ENDOGLUCANASE V CELLOHEXAOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOHEXAOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4V15
 
 | | Crystal structure of D-threonine aldolase from Alcaligenes xylosoxidans | | Descriptor: | D-THREONINE ALDOLASE, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Uhl, M.K, Oberdorfer, G, Steinkellner, G, Riegler, L, Schuermann, M, Gruber, K. | | Deposit date: | 2014-09-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of D-Threonine Aldolase from Alcaligenes Xylosoxidans Provides Insight Into a Metal Ion Assisted Plp-Dependent Mechanism.
Plos One, 10, 2015
|
|
3ENG
 
 | | STRUCTURE OF ENDOGLUCANASE V CELLOBIOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOBIOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1DYM
 
 | | Humicola insolens Endocellulase Cel7B (EG 1) E197A Mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Moraz, O, Driguez, H, Schulein, M. | | Deposit date: | 2000-02-03 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335 ( Pt 2), 1998
|
|
4RMG
 
 | | Human Sirt2 in complex with SirReal2 and NAD+ | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, NAD-dependent protein deacetylase sirtuin-2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RMI
 
 | | Human Sirt2 in complex with SirReal1 and Ac-Lys-OTC peptide | | Descriptor: | Ac-Lys-OTC peptide, N-(5-benzyl-1,3-thiazol-2-yl)-2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetamide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RMJ
 
 | | Human Sirt2 in complex with ADP ribose and nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
