8OIT
 
 | | 39S human mitochondrial large ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 16S rRNA, 39S ribosomal protein L1, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
8OIQ
 
 | | 39S mammalian mitochondrial large ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 16S rRNA, 39S ribosomal protein L1, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
4NY6
 
 | | Neutron structure of leucine and valine methyl protonated type III antifreeze | | Descriptor: | Type-3 ice-structuring protein HPLC 12 | | Authors: | Fisher, S.J, Blakeley, M.P, Howard, E.I, Petite-Haertlein, I, Haertlein, M, Mitschler, A, Cousido-Siah, A, Salvaya, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A.D. | | Deposit date: | 2013-12-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.05 Å), X-RAY DIFFRACTION | | Cite: | Perdeuteration: improved visualization of solvent structure in neutron macromolecular crystallography.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2XTI
 
 | | Asparaginyl-tRNA synthetase from Brugia malayi complexed with ATP:Mg and L-Asp-beta-NOH adenylate:PPi:Mg | | Descriptor: | 5'-O-[(R)-{[(2S)-2-amino-4-(hydroxyamino)-4-oxobutanoyl]oxy}(hydroxy)phosphoryl]adenosine, ADENOSINE-5'-TRIPHOSPHATE, ASPARAGINYL-TRNA SYNTHETASE, ... | | Authors: | Crepin, T, Haertlein, M, Kron, M, Cusack, S. | | Deposit date: | 2010-10-10 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Hybrid Structural Model of the Complete Brugia Malayi Cytoplasmic Asparaginyl-tRNA Synthetase.
J.Mol.Biol., 405, 2011
|
|
8OIP
 
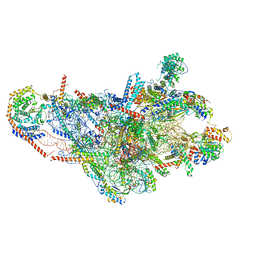 | | 28S mammalian mitochondrial small ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 12S rRNA, 28S ribosomal protein S15, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
8OIR
 
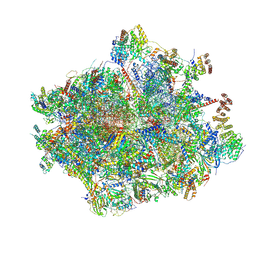 | | 55S human mitochondrial ribosome with mtRF1 and P-site tRNA | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
8OIS
 
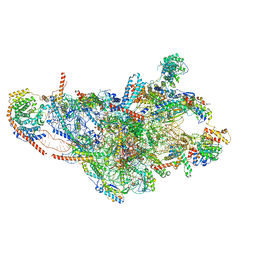 | | 28S human mitochondrial small ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 12S rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
5LFQ
 
 | | Crystal Structure of the Bacterial Proteasome Activator Bpa of Mycobacterium tuberculosis (space group P3) | | Descriptor: | Bacterial proteasome activator | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
1OF9
 
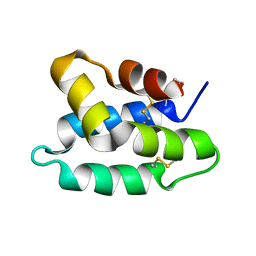 | | Solution structure of the pore forming toxin of entamoeba histolytica (Amoebapore A) | | Descriptor: | PORE-FORMING PEPTIDE AMEOBAPORE A | | Authors: | Hecht, O, Schleinkofer, K, Bruhn, H, Leippe, M, Van Nuland, N, Dingley, A.J, Grotzinger, J. | | Deposit date: | 2003-04-09 | | Release date: | 2004-02-26 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pore-Forming Protein of Entamoeba Histolytica
J.Biol.Chem., 279, 2004
|
|
1A3H
 
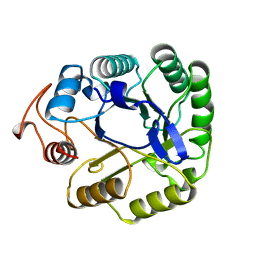 | |
8PPL
 
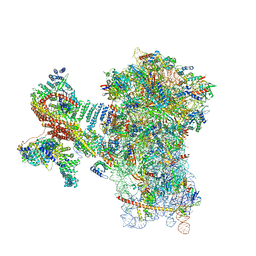 | | MERS-CoV Nsp1 bound to the human 43S pre-initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
5MMM
 
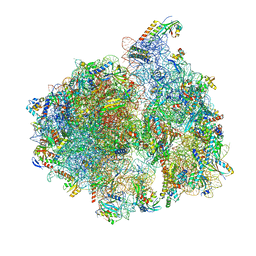 | | Structure of the 70S chloroplast ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein 2, ... | | Authors: | Bieri, P, Leibundgut, M, Saurer, M, Boehringer, D, Ban, N. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The complete structure of the chloroplast 70S ribosome in complex with translation factor pY.
EMBO J., 36, 2017
|
|
1HCD
 
 | | STRUCTURE OF HISACTOPHILIN IS SIMILAR TO INTERLEUKIN-1 BETA AND FIBROBLAST GROWTH FACTOR | | Descriptor: | HISACTOPHILIN | | Authors: | Habazettl, J, Gondol, D, Wiltscheck, R, Otlewski, J, Schleicher, M, Holak, T.A. | | Deposit date: | 1994-05-03 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor.
Nature, 359, 1992
|
|
5MMI
 
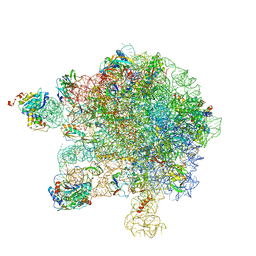 | | Structure of the large subunit of the chloroplast ribosome | | Descriptor: | 23S ribosomal RNA, 4.5S ribosomal RNA, 50S ribosomal protein 6, ... | | Authors: | Bieri, P, Leibundgut, M, Saurer, M, Boehringer, D, Ban, N. | | Deposit date: | 2016-12-10 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The complete structure of the chloroplast 70S ribosome in complex with translation factor pY.
EMBO J., 36, 2017
|
|
5MMJ
 
 | | Structure of the small subunit of the chloroplast ribosome | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein 2, chloroplastic, ... | | Authors: | Bieri, P, Leibundgut, M, Saurer, M, Boehringer, D, Ban, N. | | Deposit date: | 2016-12-10 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The complete structure of the chloroplast 70S ribosome in complex with translation factor pY.
EMBO J., 36, 2017
|
|
4A3H
 
 | | 2',4' DINITROPHENYL-2-DEOXY-2-FLURO-B-D-CELLOBIOSIDE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS AT 1.6 A RESOLUTION | | Descriptor: | 2,4-DINITROPHENYL-2-DEOXY-2-FLUORO-BETA-D-CELLOBIOSIDE, PROTEIN (ENDOGLUCANASE) | | Authors: | Davies, G.J, Brzozowski, A.M, Andersen, K, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-22 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
1HCE
 
 | | STRUCTURE OF HISACTOPHILIN IS SIMILAR TO INTERLEUKIN-1 BETA AND FIBROBLAST GROWTH FACTOR | | Descriptor: | HISACTOPHILIN | | Authors: | Habazettl, J, Gondol, D, Wiltscheck, R, Otlewski, J, Schleicher, M, Holak, T.A. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor.
Nature, 359, 1992
|
|
5LRT
 
 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP and Phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Depupylase, ... | | Authors: | Bolten, M, Vahlensieck, C, Lipp, C, Leibundgut, M, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-08-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Depupylase Dop Requires Inorganic Phosphate in the Active Site for Catalysis.
J. Biol. Chem., 292, 2017
|
|
8PPK
 
 | | Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
2OWI
 
 | | Solution structure of the RGS domain from human RGS18 | | Descriptor: | Regulator of G-protein signaling 18 | | Authors: | Higman, V.A, Leidert, M, Bray, J, Elkins, J, Soundararajan, M, Doyle, D.A, Gileadi, C, Phillips, C, Schoch, G, Yang, X, Brockmann, C, Schmieder, P, Diehl, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-16 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7P6M
 
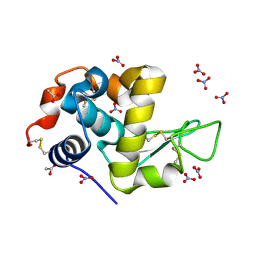 | | Hydrogenated refolded hen egg-white lysozyme | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | The impact of folding modes and deuteration on the atomic resolution structure of hen egg-white lysozyme.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5MPP
 
 | | Structure of AaLS-wt | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Sasaki, E, Boehringer, D, Leibundgut, M, Ban, N, Hilvert, D. | | Deposit date: | 2016-12-17 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and assembly of scalable porous protein cages.
Nat Commun, 8, 2017
|
|
5MQ7
 
 | | Structure of AaLS-13 | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Sasaki, E, Boehringer, D, Leibundgut, M, Ban, N, Hilvert, D. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structure and assembly of scalable porous protein cages.
Nat Commun, 8, 2017
|
|
1DYM
 
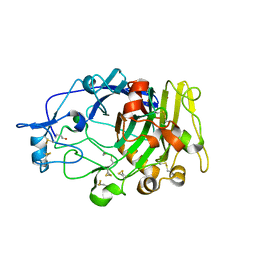 | | Humicola insolens Endocellulase Cel7B (EG 1) E197A Mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Moraz, O, Driguez, H, Schulein, M. | | Deposit date: | 2000-02-03 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335 ( Pt 2), 1998
|
|
5MQ3
 
 | | Structure of AaLS-neg | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Sasaki, E, Boehringer, D, Leibundgut, M, Ban, N, Hilvert, D. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure and assembly of scalable porous protein cages.
Nat Commun, 8, 2017
|
|
