6YR7
 
 | | 14-3-3 sigma in complex with hDMX-342+367 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein Mdm4 | | Authors: | Wolter, M, Srdanovic, S, Warriner, S, Wilson, A, Ottmann, C. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
4UDE
 
 | | An oligomerization domain confers pioneer properties to the LEAFY master floral regulator | | Descriptor: | GINLFY PROTEIN, GLYCEROL, TETRAETHYLENE GLYCOL | | Authors: | Nanao, M.H, Sayou, C, Dumas, R, Parcy, F. | | Deposit date: | 2014-12-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Sam Oligomerization Domain Shapes the Genomic Binding Landscape of the Leafy Transcription Factor
Nat.Commun., 7, 2016
|
|
6KZJ
 
 | | Crystal structure of Ankyrin B/NdeL1 complex | | Descriptor: | Ankyrin-2, Nuclear distribution protein nudE-like 1 | | Authors: | Ye, J, Li, J, Ye, F, Zhang, M, Zhang, Y, Wang, C. | | Deposit date: | 2019-09-24 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic insights into the interactions of dynein regulator Ndel1 with neuronal ankyrins and implications in polarity maintenance.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YRR
 
 | | Heimdallarchaeota profilin | | Descriptor: | Profilin | | Authors: | Celestine, C. | | Deposit date: | 2020-04-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Heimdallarchaea encodes profilin with eukaryotic-like actin regulation and polyproline binding
To Be Published
|
|
4UCW
 
 | | Structure of the T18V small subunit mutant of D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Abou-Hamdan, A, Ceccaldi, P, Lebrette, H, Guttierez-Sanz, O, Richaud, P, Cournac, L, Guigliarelli, B, deLacey, A.L, Leger, C, Volbeda, A, Burlat, B, Dementin, S. | | Deposit date: | 2014-12-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Threonine Stabilizes the Nic and Nir Catalytic Intermediates of [Nife]-Hydrogenase.
J.Biol.Chem., 290, 2015
|
|
6L47
 
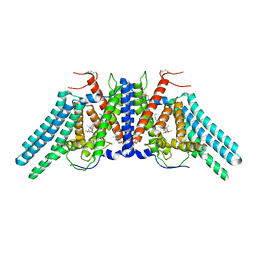 | | Structure of the human sterol O-acyltransferase 1 in complex with CI-976 | | Descriptor: | 2,2-dimethyl-N-(2,4,6-trimethoxyphenyl)dodecanamide, CHOLESTEROL, Sterol O-acyltransferase 1 | | Authors: | Chen, L, Guan, C, Niu, Y. | | Deposit date: | 2019-10-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor.
Nat Commun, 11, 2020
|
|
4UF8
 
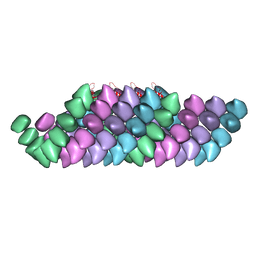 | | Electron cryo-microscopy structure of PB1-p62 filaments | | Descriptor: | SEQUESTOSOME-1 | | Authors: | Ciuffa, R, Lamark, T, Tarafder, A, Guesdon, A, Rybina, S, Hagen, W.J.H, Johansen, T, Sachse, C. | | Deposit date: | 2015-03-15 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | The Selective Autophagy Receptor P62 Forms a Flexible Filamentous Helical Scaffold.
Cell Rep., 11, 2015
|
|
6YU6
 
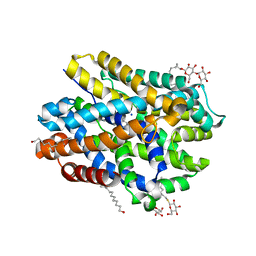 | | Crystal structure of MhsT in complex with L-leucine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, LEUCINE, SODIUM ION, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU4
 
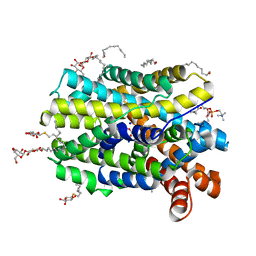 | | Crystal structure of MhsT in complex with L-4F-phenylalanine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4-FLUORO-L-PHENYLALANINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
4UJD
 
 | | mammalian 80S HCV-IRES initiation complex with eIF5B PRE-like state | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Eif5B on Hcv Ires
Nat.Struct.Mol.Biol., 21, 2014
|
|
6YWC
 
 | | De novo designed protein 4E1H_95 in complex with 101F antibody | | Descriptor: | Antibody 101F, Heavy Chain, light chain, ... | | Authors: | Yang, C, Sesterhenn, F, Pojer, F, Correia, B.E. | | Deposit date: | 2020-04-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bottom-up de novo design of functional proteins with complex structural features.
Nat.Chem.Biol., 17, 2021
|
|
4UHW
 
 | | Human aldehyde oxidase | | Descriptor: | ALDEHYDE OXIDASE, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Coelho, C, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2015-03-26 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Xenobiotic and Inhibitor Binding to Human Aldehyde Oxidase
Nat.Chem.Biol., 11, 2015
|
|
6LDH
 
 | |
6LDP
 
 | | Structure of CDK5R1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Cyclin-dependent kinase 5 activator 1, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-22 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
4UMN
 
 | | Structure of a stapled peptide antagonist bound to Nutlin-resistant Mdm2. | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, M06 | | Authors: | Chee, S, Wongsantichon, J, Quah, S, Robinson, R.C, Verma, C, Lane, D.P, Brown, C.J, Ghadessy, F.J. | | Deposit date: | 2014-05-20 | | Release date: | 2014-05-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of a stapled peptide antagonist bound to nutlin-resistant Mdm2.
PLoS ONE, 9, 2014
|
|
6Z27
 
 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV LCP crystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A.G, Fufina, T.Y, Vasilieva, L.G, Betzel, C, Selikhanov, G.K. | | Deposit date: | 2020-05-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel approaches for the lipid sponge phase crystallization of the Rhodobacter sphaeroides photosynthetic reaction center.
Iucrj, 7, 2020
|
|
4UBW
 
 | |
4UN3
 
 | | Crystal structure of Cas9 bound to PAM-containing DNA target | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
6Z4X
 
 | | Structure of the CAK complex form Chaetomium thermophilum bound to ATP-gamma-S | | Descriptor: | CHLORIDE ION, CYCLIN domain-containing protein, MAGNESIUM ION, ... | | Authors: | Peissert, S, Kuper, J, Kisker, C. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for CDK7 activation by MAT1 and Cyclin H.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KWC
 
 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
6KWK
 
 | | Crystal structure of pSLA-1*0401 complex with FMDV-derived epitope MTAHITVPY | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
4UCT
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-amino-6-methyl-5-(propan-2-yloxy)-3H-[1,2,4]triazolo[1,5-a]pyrimidin-8-ium, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCX
 
 | | Structure of the T18G small subunit mutant of D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Abou-Hamdan, A, Ceccaldi, P, Lebrette, H, Guttierez-Sanz, O, Richaud, P, Cournac, L, Guigliarelli, B, deLacey, A.L, Leger, C, Volbeda, A, Burlat, B, Dementin, S. | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A threonine stabilizes the NiC and NiR catalytic intermediates of [NiFe]-hydrogenase.
J. Biol. Chem., 290, 2015
|
|
4UDV
 
 | | Cryo-EM structure of TMV at 3.35 A resolution | | Descriptor: | 5'-D(*GP*AP*AP)-3', CAPSID PROTEIN | | Authors: | Fromm, S.A, Bharat, T.A.M, Jakobi, A.J, Hagen, W.J.H, Sachse, C. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Seeing Tobacco Mosaic Virus Through Direct Electron Detectors.
J.Struct.Biol., 189, 2015
|
|
4XCU
 
 | | Crystal Structure of FGFR4 with an Irreversible Inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-(2-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)quinazolin-2-yl]amino}-3-methylphenyl)propanamide, SULFATE ION | | Authors: | Kim, J.L, Miduturu, C, Hodous, B, Brooijmans, N, Bifulco, N, Guzi, T. | | Deposit date: | 2014-12-18 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | First Selective Small Molecule Inhibitor of FGFR4 for the Treatment of Hepatocellular Carcinomas with an Activated FGFR4 Signaling Pathway.
Cancer Discov, 5, 2015
|
|
