8TII
 
 | |
8TIN
 
 | |
8UCX
 
 | |
4NWJ
 
 | | Crystal structure of phosphopglycerate mutase from Staphylococcus aureus in 3-phosphoglyceric acid bound form. | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, 3-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION | | Authors: | Roychowdhury, A, Bose, M, Kundu, A, Gujar, A, Das, A.K. | | Deposit date: | 2013-12-06 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Complete catalytic cycle of cofactor-independent phosphoglycerate mutase involves a spring-loaded mechanism
Febs J., 282, 2015
|
|
4NWX
 
 | | Crystal structure of phosphoglycerate mutase from Staphylococcus aureus in 2-phosphoglyceric acid bound form | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, 2-PHOSPHOGLYCERIC ACID, ... | | Authors: | Roychowdhury, A, Kundu, A, Bose, M, Gujar, A, Das, A.K. | | Deposit date: | 2013-12-07 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Complete catalytic cycle of cofactor-independent phosphoglycerate mutase involves a spring-loaded mechanism
Febs J., 282, 2015
|
|
4JYT
 
 | | Crystal Structure of Matriptase in complex with Inhibitor | | Descriptor: | 4,4'-[{3-[(naphthalen-2-ylsulfonyl)amino]pyridine-2,6-diyl}bis(oxy)]dibenzenecarboximidamide, Suppressor of tumorigenicity 14 protein | | Authors: | Subramanya, H.S, Ravi, B.C, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S. | | Deposit date: | 2013-04-01 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
ACS MED.CHEM.LETT., 4, 2013
|
|
4JZI
 
 | | Crystal Structure of Matriptase in complex with Inhibitor". | | Descriptor: | N-(trans-4-aminocyclohexyl)-2,6-bis(4-carbamimidoylphenoxy)pyridine-4-carboxamide, Suppressor of tumorigenicity 14 protein | | Authors: | Subramanya, H.S, Chandra, R.B, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S. | | Deposit date: | 2013-04-03 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
ACS MED.CHEM.LETT., 4, 2013
|
|
4JZ1
 
 | | Crystal Structure of Matriptase in complex with Inhibitor | | Descriptor: | 4,4'-[(3-{[(4-fluorophenyl)sulfonyl]amino}pyridine-2,6-diyl)bis(oxy)]dibenzenecarboximidamide, Suppressor of tumorigenicity 14 protein | | Authors: | Subramanya, H.S, Ravi, B.C, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S. | | Deposit date: | 2013-04-02 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
ACS MED.CHEM.LETT., 4, 2013
|
|
4MY4
 
 | | Crystal structure of phosphoglycerate mutase from Staphylococcus aureus. | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, MANGANESE (II) ION | | Authors: | Roychowdhury, A, Kundu, A, Gujar, A, Bose, M, Das, A.K. | | Deposit date: | 2013-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complete catalytic cycle of cofactor-independent phosphoglycerate mutase involves a spring-loaded mechanism
Febs J., 282, 2015
|
|
4QAX
 
 | | Crystal structure of post-catalytic binary complex of Phosphoglycerate mutase from Staphylococcus aureus | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, 2-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION | | Authors: | Roychowdhury, A, Kundu, A, Bose, M, Gujar, A, Das, A.K. | | Deposit date: | 2014-05-06 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | STRUCTURAL AND FUNCCTIONAL ANALYSIS of PHOSPHOGLYCERATE MUTASE from STAPHYLOCOCCUS AUREUS
To be Published
|
|
4RVC
 
 | |
3M1L
 
 | | Crystal structure of a C-terminal trunacted mutant of a putative ketoacyl reductase (FabG4) from Mycobacterium tuberculosis H37Rv at 2.5 Angstrom resolution | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) reductase, ACETATE ION | | Authors: | Dutta, D, Bhattacharyya, S, Saha, B, Das, A.K. | | Deposit date: | 2010-03-05 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of FabG4 from Mycobacterium tuberculosis reveals the importance of C-terminal residues in ketoreductase activity
J.Struct.Biol., 174, 2011
|
|
8VJ9
 
 | |
8VZ4
 
 | |
8DS9
 
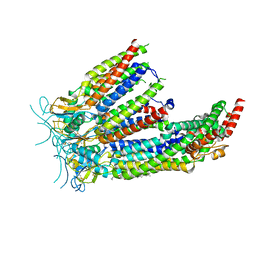 | | LRRC8A:C in MSPE3D1 nanodisc top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DR8
 
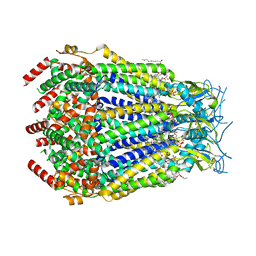 | | LRRC8A:C conformation 2 (oblong) top mask | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DRO
 
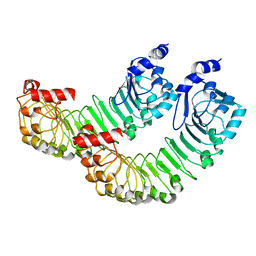 | | LRRC8A:C conformation 1 (round) LRR focus 2 | | Descriptor: | Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562 | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DRA
 
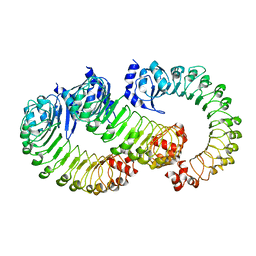 | | LRRC8A:C conformation 2 (oblong) LRR mask | | Descriptor: | Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DSA
 
 | | LRRC8A:C in MSP1E3D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DRN
 
 | | LRRC8A:C conformation 1 (round) LRR focus 1 | | Descriptor: | Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DRQ
 
 | | LRRC8A:C conformation 1 (round) LRR focus 3 | | Descriptor: | Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562 | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DRE
 
 | | LRRC8A:C conformation 2 (oblong) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DRK
 
 | | LRRC8A:C conformation 1 (round) top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DS3
 
 | | LRRC8A:C conformation 1 (round) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8F7D
 
 | | LRRC8A(T48D):C conformation 2 top focus | | Descriptor: | Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
