5UE6
 
 | | Structure of nitrite reductase AniA from Neisseria gonorrhoeae, space group I4122 | | Descriptor: | COPPER (II) ION, Nitrite reductase, SODIUM ION | | Authors: | Hamza, A, Williamson, Z.A, Reed, R.W, Sikora, A.E, Korotkov, K.V. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Peptide Inhibitors Targeting the Neisseria gonorrhoeae Pivotal Anaerobic Respiration Factor AniA.
Antimicrob. Agents Chemother., 61, 2017
|
|
1P4N
 
 | | Crystal Structure of Weissella viridescens FemX:UDP-MurNAc-pentapeptide complex | | Descriptor: | FemX, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Biarrotte-Sorin, S, Maillard, A, Delettre, J, Sougakoff, W, Arthur, M, Mayer, C. | | Deposit date: | 2003-04-23 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Weissella viridescens FemX and its complex with UDP-MurNAc-pentapeptide: insights into FemABX family substrates recognition.
Structure, 12, 2004
|
|
5TGF
 
 | | Crystal structure of putative beta-lactamase from Bacteroides dorei DSM 17855 | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein | | Authors: | Nocek, B, Hatzos-Skintges, C, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-27 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of a beta-lactamase from Bacteroides dorei DSM 17855
To Be Published
|
|
3UUD
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with estradiol | | Descriptor: | 1,2-ETHANEDIOL, ESTRADIOL, Estrogen receptor, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2011-11-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UUA
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with bisphenol-AF | | Descriptor: | 4,4'-(1,1,1,3,3,3-hexafluoropropane-2,2-diyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 1 | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2011-11-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UU7
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with bisphenol-A | | Descriptor: | 4,4'-PROPANE-2,2-DIYLDIPHENOL, Estrogen receptor, Nuclear receptor coactivator 1 | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2011-11-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UUC
 
 | | Crystal structure of hERa-LBD (wt) in complex with bisphenol-C | | Descriptor: | 4,4'-(2,2-dichloroethene-1,1-diyl)diphenol, Estrogen receptor | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2011-11-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8CBH
 
 | | SHP2 in complex with a novel allosteric inhibitor | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11, [(1~{S},6~{R},7~{S})-3-[3-[2,3-bis(chloranyl)phenyl]-2~{H}-pyrazolo[3,4-b]pyrazin-6-yl]-7-(4-methyl-1,3-thiazol-2-yl)-3-azabicyclo[4.1.0]heptan-7-yl]methanamine | | Authors: | di Fabio, R, Petrocchi, A. | | Deposit date: | 2023-01-25 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of a Novel Series of Imidazopyrazine Derivatives as Potent SHP2 Allosteric Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
5UDI
 
 | | IFIT1 monomeric mutant (L457E/L464E) with m7Gppp-AAAA (syn and anti conformations of cap) | | Descriptor: | CALCIUM ION, Interferon-induced protein with tetratricopeptide repeats 1, RNA (5'-D(*(GTA))-R(P*AP*AP*A)-3'), ... | | Authors: | Abbas, Y.M, Martinez-Montero, S, Damha, M.J, Nagar, B. | | Deposit date: | 2016-12-27 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2'-O methylations.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UDJ
 
 | | IFIT1 monomeric mutant (L457E/L464E) with Gppp-AAAA | | Descriptor: | CALCIUM ION, Interferon-induced protein with tetratricopeptide repeats 1, RNA (5'-D(*(G3A))-R(P*AP*AP*A)-3'), ... | | Authors: | Abbas, Y.M, Martinez-Montero, S, Damha, M.J, Nagar, B. | | Deposit date: | 2016-12-27 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2'-O methylations.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7K2Z
 
 | | Crystal structure of Pisum sativum KAI2 Apo form | | Descriptor: | GLYCEROL, PsKAI2B protein | | Authors: | Guercio, A.M, Shabek, N. | | Deposit date: | 2020-09-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and functional analyses explain Pea KAI2 receptor diversity and reveal stereoselective catalysis during signal perception.
Commun Biol, 5, 2022
|
|
7K38
 
 | |
5UDL
 
 | | IFIT1 N216A monomeric mutant (L457E/L464E) with m7Gppp-AAAA (anti conformation of cap) | | Descriptor: | CALCIUM ION, Interferon-induced protein with tetratricopeptide repeats 1, RNA (5'-D(*(GTA))-R(P*AP*AP*A)-3'), ... | | Authors: | Abbas, Y.M, Martinez-Montero, S, Damha, M.J, Nagar, B. | | Deposit date: | 2016-12-27 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2'-O methylations.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UDK
 
 | | IFIT1 monomeric mutant (L457E/L464E) with PPP-AAAA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Abbas, Y.M, Martinez-Montero, S, Damha, M.J, Nagar, B. | | Deposit date: | 2016-12-27 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2'-O methylations.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8Q8J
 
 | | Crystal structure of human GPX4-R152H | | Descriptor: | Glutathione peroxidase | | Authors: | Napolitano, V, Mourao, A, Kolonko, M, Bostock, M.J, Sattler, M, Conrad, M, Popowicz, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human GPX4-R152H
To Be Published
|
|
8Q8N
 
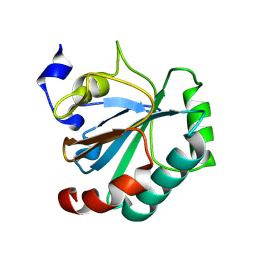 | | Crystal structure of human GPX4-U46C-I129S-L130S | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Bostock, M.J, Mourao, A, Napolitano, V, Sattler, M, Conrad, M, Popowicz, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An ultra-rare variant of GPX4 reveals the structural basis to avert neurodegeneration
To Be Published
|
|
8R8M
 
 | |
8R6C
 
 | |
3QYW
 
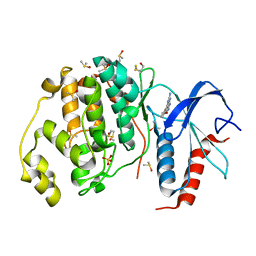 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 6-(3-bromophenyl)-7H-purin-2-amine, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Gelin, M, Pochet, S, Hoh, F, Pirochi, M, Guichou, J.-F, Ferrer, J.-L, Labesse, G. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5N6I
 
 | | Crystal structure of mouse cGAS in complex with 39 bp DNA | | Descriptor: | Cyclic GMP-AMP synthase, DNA (36-MER), DNA (37-MER), ... | | Authors: | Andreeva, L, Kostrewa, D, Hopfner, K.-P. | | Deposit date: | 2017-02-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders.
Nature, 549, 2017
|
|
3QYU
 
 | | Crystal structure of human cyclophilin D at 1.54 A resolution at room temperature | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F | | Authors: | Colliandre, L, Gelin, M, Labesse, G, Guichou, J.-F. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3QYZ
 
 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 5'-azido-8-bromo-5'-deoxyadenosine, BETA-MERCAPTOETHANOL, DIMETHYL SULFOXIDE, ... | | Authors: | Gelin, M, Pochet, S, Hoh, F, Pirochi, M, Guichou, J.-F, Ferrer, J.-L, Labesse, G. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4K22
 
 | | Structure of the C-terminal truncated form of E.Coli C5-hydroxylase UBII involved in ubiquinone (Q8) biosynthesis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pecqueur, L, Lombard, M, Golinelli-pimpaneau, B, Fontecave, M. | | Deposit date: | 2013-04-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ubiI, a New Gene in Escherichia coli Coenzyme Q Biosynthesis, Is Involved in Aerobic C5-hydroxylation.
J.Biol.Chem., 288, 2013
|
|
4DK9
 
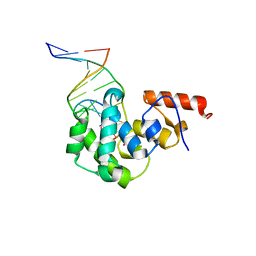 | | Crystal Structure of MBD4 Catalytic Domain Bound to Abasic DNA | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*GP*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*T)-3', Methyl-CpG-binding domain protein 4, ... | | Authors: | Manvilla, B.A, Toth, E.A, Drohat, A.C. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal Structure of Human Methyl-Binding Domain IV Glycosylase Bound to Abasic DNA.
J.Mol.Biol., 420, 2012
|
|
8R8A
 
 | |
