5WB6
 
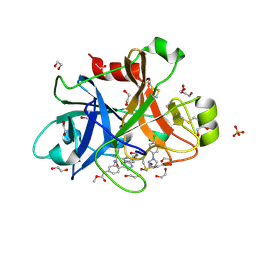 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(11S)-11-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-6-fluoro-2-oxo-1,3,4,10,11,13-hexahydro-2H-5,9:15,12-di(azeno)-1,13-benzodiazacycloheptadecin-18-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrocyclic factor XIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5WSN
 
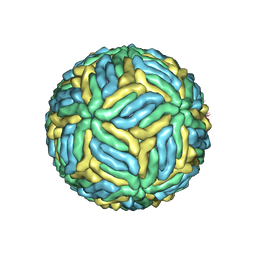 | | Structure of Japanese encephalitis virus | | Descriptor: | E protein, M protein | | Authors: | Wang, X, Zhu, L, Li, S, Yuan, S, Qin, C, Fry, E.E, Stuart, I.D, Rao, Z. | | Deposit date: | 2016-12-07 | | Release date: | 2017-05-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability
Nat Commun, 8, 2017
|
|
2P9C
 
 | |
2PA3
 
 | |
2P9G
 
 | |
2P9E
 
 | |
3OEV
 
 | | Structure of yeast 20S open-gate proteasome with Compound 25 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(benzyloxy)-N-[(2S,3R)-3-hydroxy-1-{[(2S)-1-{[(3-methylthiophen-2-yl)methyl]amino}-1-oxo-4-phenylbutan-2-yl]amino}-1-oxobutan-2-yl]benzamide, MAGNESIUM ION, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-08-13 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OEU
 
 | | Structure of yeast 20S open-gate proteasome with Compound 24 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-{(2S)-1-[(2-chlorobenzyl)amino]-1-oxo-4-phenylbutan-2-yl}-N~2~-[3-(2-methylphenyl)propanoyl]-L-threoninamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-08-13 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3SDI
 
 | | Structure of yeast 20S open-gate proteasome with Compound 20 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N~4~-(2,2-dimethylpropyl)-N~1~-{(2S)-1-[(4-methylbenzyl)amino]-1-oxo-4-phenylbutan-2-yl}-N~2~-[(5-methyl-1,2-oxazol-3-yl)carbonyl]-L-aspartamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2011-06-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3SDK
 
 | | Structure of yeast 20S open-gate proteasome with Compound 34 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-[(2S)-3-(3-tert-butyl-1,2,4-oxadiazol-5-yl)-1-({(2S)-1-[(4-methylbenzyl)amino]-1-oxo-4-phenylbutan-2-yl}amino)-1-oxopropan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2011-06-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3U3I
 
 | | A RNA binding protein from Crimean-Congo hemorrhagic fever virus | | Descriptor: | Nucleocapsid protein | | Authors: | Guo, Y, Wang, W.M, Ji, W, Deng, M, Sun, Y.N, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2011-10-06 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | Descriptor: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | Authors: | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | Descriptor: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
4LPG
 
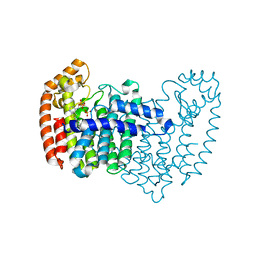 | | Crystal structure of human FPPS in complex with CL01131 | | Descriptor: | ({[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}methanediyl)bis(phosphonic acid), Farnesyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-07-16 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Multistage screening reveals chameleon ligands of the human farnesyl pyrophosphate synthase: implications to drug discovery for neurodegenerative diseases.
J.Med.Chem., 57, 2014
|
|
8D36
 
 | |
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | Descriptor: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | Authors: | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
4LPH
 
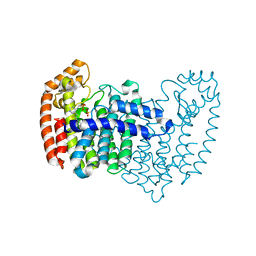 | | Crystal structure of human FPPS in complex with CL03093 | | Descriptor: | ({[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}methyl)phosphonic acid, Farnesyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-07-16 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multistage screening reveals chameleon ligands of the human farnesyl pyrophosphate synthase: implications to drug discovery for neurodegenerative diseases.
J.Med.Chem., 57, 2014
|
|
5YUD
 
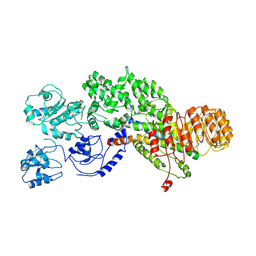 | | Flagellin derivative in complex with the NLR protein NAIP5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Baculoviral IAP repeat-containing protein 1e, Phase 2 flagellin,Flagellin | | Authors: | Yang, X.R, Yang, F, Wang, W.G, Lin, G.Z. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structural basis for specific flagellin recognition by the NLR protein NAIP5.
Cell Res., 28, 2018
|
|
8XJJ
 
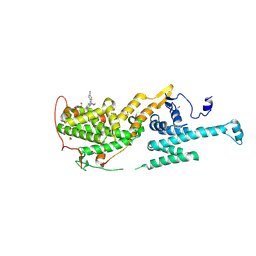 | | Co-crystal structure of SOS-1 and a potent, selective and orally bioavailable SOS1 inhibitor RGT-018 | | Descriptor: | 1,2-ETHANEDIOL, 5-[4-[[(1~{R})-1-[3-[bis(fluoranyl)methyl]-2-fluoranyl-phenyl]ethyl]amino]-2-methyl-6-morpholin-4-yl-7-oxidanylidene-pyrido[4,3-d]pyrimidin-8-yl]pyridine-2-carbonitrile, Son of sevenless homolog 1 | | Authors: | Ren, X. | | Deposit date: | 2023-12-21 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of RGT-018: a Potent, Selective and Orally Bioavailable SOS1 Inhibitor for KRAS-driven Cancers.
Mol.Cancer Ther., 2024
|
|
5O8W
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE YEAST ELONGATION FACTOR COMPLEX EEF1A:EEF1BA | | Descriptor: | Elongation factor 1-alpha, Elongation factor 1-beta, GLUTAMINE, ... | | Authors: | Wirth, C, Andersen, G.R, Hunte, C. | | Deposit date: | 2017-06-14 | | Release date: | 2017-08-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Protein glutaminylation is a yeast-specific posttranslational modification of elongation factor 1A.
J. Biol. Chem., 292, 2017
|
|
7LQD
 
 | | Structure of Human MPS1 (TTK) covalently bound to RMS-07 inhibitor | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, Dual specificity protein kinase TTK or monopolar spindle 1 kinase, GLYCEROL, ... | | Authors: | Santiago, A.S, dos Reis, C.V, Serafim, R.A.M, Massirer, K.B, Arruda, P, Edwards, A.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-02-13 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of the First Covalent Monopolar Spindle Kinase 1 (MPS1/TTK) Inhibitor.
J.Med.Chem., 65, 2022
|
|
7CKZ
 
 | | Cryo-EM structure of Dopamine and LY3154207 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKY
 
 | | Cryo-EM structure of PW0464 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | 6-[4-[3-[bis(fluoranyl)methoxy]pyridin-2-yl]oxy-2-methyl-phenyl]-1,5-dimethyl-pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
