6UJ8
 
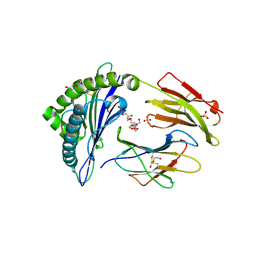 | | Crystal structure of HLA-B*07:02 with wild-type IDH2 peptide | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, HLA class I histocompatibility antigen, ... | | Authors: | Miller, M.S, Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
6UJ9
 
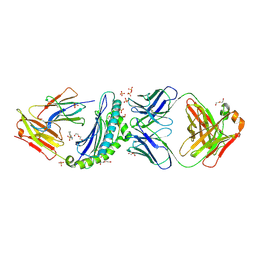 | | Crystal structure of HLA-B*07:02 with R140Q mutant IDH2 peptide in complex with Fab | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, M.S, Thirawatananond, P, Aytenfisu, T.Y, Wright, K, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
6UJ7
 
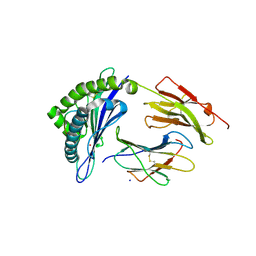 | | Crystal structure of HLA-B*07:02 with R140Q mutant IDH2 peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Miller, M.S, Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
7MYG
 
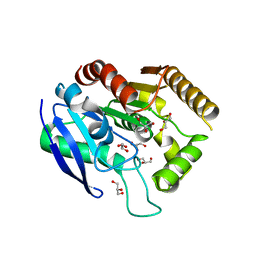 | | M. tb Ag85C modified by THL-10d | | Descriptor: | Diacylglycerol acyltransferase, GLYCEROL | | Authors: | Sudasinghe, T.D, Ronning, D.R. | | Deposit date: | 2021-05-21 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Total Synthesis of Tetrahydrolipstatin, Its Derivatives, and Evaluation of Their Ability to Potentiate Multiple Antibiotic Classes against Mycobacterium Species.
Acs Infect Dis., 7, 2021
|
|
7MZT
 
 | |
7NA2
 
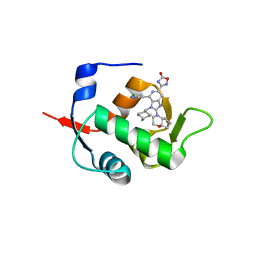 | | HDM2 in complex with compound 56 | | Descriptor: | 3-[4-(5-chloropyridin-3-yl)-2-[(4aR,7aR)-hexahydrocyclopenta[b][1,4]oxazin-4(4aH)-yl]-3-{[(1r,4R)-4-methylcyclohexyl]methyl}-3H-imidazo[4,5-c]pyridin-6-yl]-1,2,4-oxadiazol-5(4H)-one, Isoform 11 of E3 ubiquitin-protein ligase Mdm2 | | Authors: | Scapin, G. | | Deposit date: | 2021-06-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7NA3
 
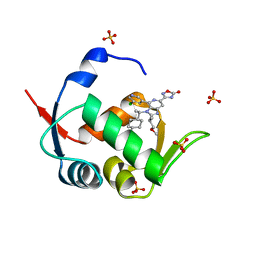 | | HDM2 in complex with compound 62 | | Descriptor: | 3-[4-(5-chloropyridin-3-yl)-2-[(2S)-1-methoxypropan-2-yl]-3-{(1R)-1-[(1r,4R)-4-methylcyclohexyl]ethyl}-3H-imidazo[4,5-c]pyridin-6-yl]-1,2,4-oxadiazol-5(4H)-one, Isoform 11 of E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Scapin, G. | | Deposit date: | 2021-06-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7NA1
 
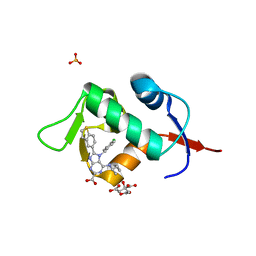 | | HDM2 in complex with compound 2 | | Descriptor: | 8-(1-benzothiophen-5-yl)-7-[(4-chlorophenyl)methyl]-6-{[(1R)-1-cyclopropylethyl]amino}-7H-purine-2-carboxylic acid, CITRIC ACID, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Scapin, G. | | Deposit date: | 2021-06-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7NA4
 
 | | HDM2 in complex with compound 63 | | Descriptor: | 3-[4-(5-chloropyridin-3-yl)-2-[(R)-cyclopropyl(ethoxy)methyl]-3-{(1R)-1-[(1r,4R)-4-methylcyclohexyl]ethyl}-3H-imidazo[4,5-c]pyridin-6-yl]-1,2,4-oxadiazol-5(4H)-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Scapin, G. | | Deposit date: | 2021-06-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
6V8I
 
 | |
7N4Z
 
 | | Complex structure of NOS4 with noscapine | | Descriptor: | NOS4, SULFATE ION, noscapine | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
7N53
 
 | | Complex structure of PAP4 with papaverine | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, GLYCEROL, PAP4, ... | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
7N54
 
 | | Complex structure of GLAU4 with glaucine | | Descriptor: | GLAU4, SULFATE ION, glaucine | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
7N4W
 
 | | Complex structure of ROTU4 with rotundine | | Descriptor: | GLYCEROL, ROTU4, SULFATE ION, ... | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
1UD1
 
 | | Crystal structure of proglycinin mutant C88S | | Descriptor: | Glycinin G1 | | Authors: | Utsumi, S, Adachi, M. | | Deposit date: | 2003-04-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures and Structural Stabilities of the Disulfide Bond-Deficient Soybean Proglycinin Mutants C12G and C88S.
J.Agric.Food Chem., 51, 2003
|
|
7OJY
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 6 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-[(5-oxidanyl-1,3,4-oxadiazol-2-yl)methyl]ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OK1
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 3 | | Descriptor: | Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, ~{N}-[(5-azanyl-1,3,4-oxadiazol-2-yl)methyl]-2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]ethanamide | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OJ6
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 1 | | Descriptor: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, ACETATE ION, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, ... | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OJP
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 1 | | Descriptor: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, MAGNESIUM ION | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OJQ
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 7 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-[(5-methyl-1,3,4-oxadiazol-2-yl)methyl]ethanamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OJW
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 93 | | Descriptor: | 2-[2-(2-ethylphenoxy)ethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-~{N}-methyl-ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OK2
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 3 | | Descriptor: | Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, SULFATE ION, ~{N}-[(5-azanyl-1,3,4-oxadiazol-2-yl)methyl]-2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]ethanamide | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7ODL
 
 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-29 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
7OD3
 
 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-28 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
6V6Z
 
 | | Crystal structure of N-[(4-methoxyphenyl)sulfonyl]-N-(4-{[(4-methoxyphenyl)sulfonyl]amino}naphthalen-1-yl)glycine bound to human Keap1 Kelch domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DITHIANE DIOL, FORMIC ACID, ... | | Authors: | Lazzara, P.R, David, B.P, Ankireddy, A, Richardson, B.G, Dye, K, Ratia, K.M, Reddy, S.P, Moore, T.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synthesis and Evaluation of Noncovalent Naphthalene-Based KEAP1-NRF2 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
