7WXZ
 
 | | Crystal structure of the recombinant protein HR121 from the S2 protein of SARS-CoV-2 | | Descriptor: | Spike protein S2' | | Authors: | Zheng, Y.T, Ouyang, S, Pang, W, Lu, Y, Zhao, Y.B. | | Deposit date: | 2022-02-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A variant-proof SARS-CoV-2 vaccine targeting HR1 domain in S2 subunit of spike protein.
Cell Res., 32, 2022
|
|
6BTZ
 
 | | Crystal structure of the PI3KC2alpha C2 domain in space group C121 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, GLYCEROL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ... | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
4JZ1
 
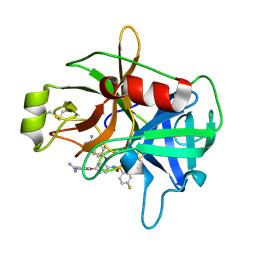 | | Crystal Structure of Matriptase in complex with Inhibitor | | Descriptor: | 4,4'-[(3-{[(4-fluorophenyl)sulfonyl]amino}pyridine-2,6-diyl)bis(oxy)]dibenzenecarboximidamide, Suppressor of tumorigenicity 14 protein | | Authors: | Subramanya, H.S, Ravi, B.C, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S. | | Deposit date: | 2013-04-02 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
ACS MED.CHEM.LETT., 4, 2013
|
|
4JYT
 
 | | Crystal Structure of Matriptase in complex with Inhibitor | | Descriptor: | 4,4'-[{3-[(naphthalen-2-ylsulfonyl)amino]pyridine-2,6-diyl}bis(oxy)]dibenzenecarboximidamide, Suppressor of tumorigenicity 14 protein | | Authors: | Subramanya, H.S, Ravi, B.C, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S. | | Deposit date: | 2013-04-01 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
ACS MED.CHEM.LETT., 4, 2013
|
|
6CVX
 
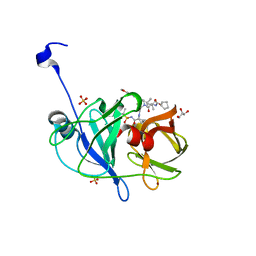 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-50 (MK-5172 linear analogue) | | Descriptor: | GLYCEROL, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-4-{[7-methoxy-3-(propan-2-yl)quinoxalin-2-yl]oxy}-L-prolinamide, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
4BL9
 
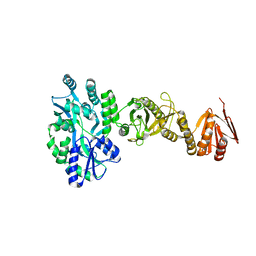 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form I) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7XI3
 
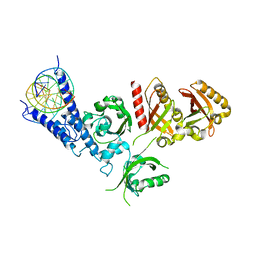 | | Crystal Structure of the NPAS4-ARNT2 heterodimer in complex with DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator 2, DNA (5'-D(P*CP*CP*AP*TP*CP*AP*CP*TP*CP*AP*CP*GP*AP*CP*CP*T)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Sun, X.N, Jing, L.Q, Li, F.W, Wu, D.L. | | Deposit date: | 2022-04-11 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.274 Å) | | Cite: | Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XI4
 
 | | Crystal Structure of the NPAS4-ARNT heterodimer in complex with DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*GP*GP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*CP*CP*AP*TP*CP*AP*CP*TP*CP*AP*CP*GP*AP*CP*CP*T)-3'), ... | | Authors: | Sun, X.N, Jing, L.Q, Li, F.W, Wu, D.L. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.707 Å) | | Cite: | Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XHV
 
 | | Crystal Structure of the NPAS4-ARNT heterodimer in complex with DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(P*CP*CP*AP*TP*CP*AP*CP*TP*CP*AP*CP*GP*AP*CP*CP*T)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Sun, X.N, Jing, L.Q, Li, F.W, Wu, D.L. | | Deposit date: | 2022-04-10 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.996 Å) | | Cite: | Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4JAI
 
 | | Crystal Structure of Aurora Kinase A in complex with N-{4-[(6-oxo-5,6-dihydrobenzo[c][1,8]naphthyridin-1-yl)amino]phenyl}benzamide | | Descriptor: | Aurora kinase A, N-{4-[(6-oxo-5,6-dihydrobenzo[c][1,8]naphthyridin-1-yl)amino]phenyl}benzamide | | Authors: | Jiang, X, Josephson, K, Huck, B, Goutopoulos, A, Karra, S. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SAR and evaluation of novel 5H-benzo[c][1,8]naphthyridin-6-one analogs as Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7V7L
 
 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
4BLD
 
 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI3p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, TRANSCRIPTIONAL ACTIVATOR GLI3, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BL8
 
 | | Crystal structure of full-length human Suppressor of fused (SUFU) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Karlstrom, M, Finta, C, Cherry, A.L, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZNX
 
 | | Crystal structure of the OTU domain of OTULIN D336A mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
3ZNZ
 
 | | Crystal structure of OTULIN OTU domain (C129A) in complex with Met1- di ubiquitin | | Descriptor: | POLYUBIQUITIN-C, PROTEIN FAM105B, SULFATE ION | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
3CPP
 
 | |
3VOZ
 
 | | Crystal structure of human glutaminase in complex with BPTES | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide), ... | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BVG
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH NATIVE ALKYLIMIDATE FORMED FROM ACETYL-LYSINE ACS2-PEPTIDE CRYSTALLIZED IN PRESENCE OF THE INHIBITOR EX-527 | | Descriptor: | (2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL ACETATE, 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ex-527 inhibits Sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
4BLA
 
 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form II) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6BU0
 
 | | Crystal structure of the PI3KC2alpha C2 domain in complex with IP6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, FORMIC ACID, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
6BUB
 
 | | Crystal structure of the PI3KC2alpha PX domain in space group P432 | | Descriptor: | GLYCEROL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, SULFATE ION | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
4BV2
 
 | | CRYSTAL STRUCTURE OF SIR2 IN COMPLEX WITH THE INHIBITOR EX-527, 2'-O-ACETYL-ADP-RIBOSE AND DEACETYLATED P53-PEPTIDE | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, CELLULAR TUMOR ANTIGEN P53, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6DKA
 
 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | CYANAMIDE, DNA damage-inducible protein, SULFATE ION, ... | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
6HHO
 
 | | Crystal structure of RIP1 kinase in complex with GSK547 | | Descriptor: | 6-[4-[(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]carbonylpiperidin-1-yl]pyrimidine-4-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer.
Cancer Cell, 34, 2018
|
|
3VOY
 
 | | Crystal structure of human glutaminase in apo form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, SULFATE ION | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
