3X2F
 
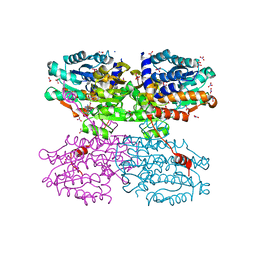 | | A Thermophilic S-Adenosylhomocysteine Hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Adenosylhomocysteinase, NITRATE ION, ... | | Authors: | Zheng, Y, Ko, T.P, Huang, C.H. | | Deposit date: | 2014-12-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structures of S-adenosylhomocysteine hydrolase from the thermophilic bacterium Thermotoga maritima.
J.Struct.Biol., 190, 2015
|
|
7YIK
 
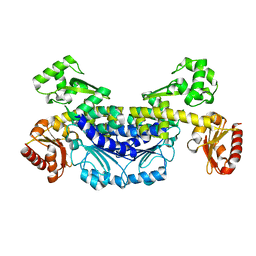 | |
7YIL
 
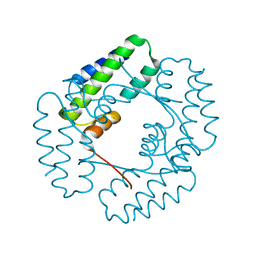 | |
3X2E
 
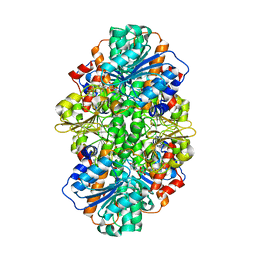 | | A thermophilic hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Adenosylhomocysteinase | | Authors: | Zheng, Y, Ko, T.P, Huang, C.H. | | Deposit date: | 2014-12-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of S-adenosylhomocysteine hydrolase from the thermophilic bacterium Thermotoga maritima.
J.Struct.Biol., 190, 2015
|
|
7YKV
 
 | | The structure of archaeal nuclease RecJ2 from Methanocaldococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, [5-(6-aminopurin-9-yl)-2-(hydroxymethyl)furan-3-yl] [5-(6-aminopurin-9-yl)-3-oxidanyl-furan-2-yl]methyl hydrogen phosphate, ... | | Authors: | Wang, W.W, Liu, X.P. | | Deposit date: | 2022-07-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The structure of archaeal nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
7YOQ
 
 | | Recj2 and CMP complex from Methanocaldococcus jannaschii | | Descriptor: | [5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)furan-2-yl]methyl dihydrogen phosphate, nuclease RecJ2 | | Authors: | Wang, W.W, Liu, X.P. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | The structure of nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
7YOS
 
 | |
7YOR
 
 | | Recj2 and dCMP complex from Methanocaldococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, [5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-furan-2-yl]methyl dihydrogen phosphate, archaeal nuclease RecJ2 | | Authors: | Wang, W.W, Liu, X.P. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structure of nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
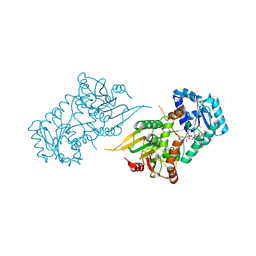
6VA7
 
 | |
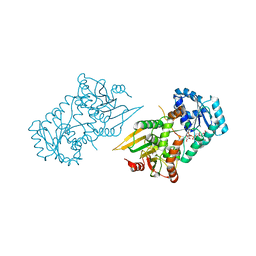
6VA8
 
 | |
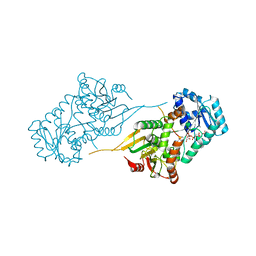
6VA9
 
 | |
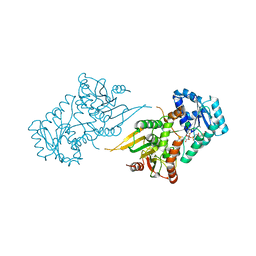
6VA0
 
 | |
6VAQ
 
 | |
7EAX
 
 | | Crystal complex of p53-V272M and antimony ion | | Descriptor: | ANTIMONY (III) ION, Cellular tumor antigen p53, ZINC ION | | Authors: | Lu, M, Tang, Y. | | Deposit date: | 2021-03-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Repurposing antiparasitic antimonials to noncovalently rescue temperature-sensitive p53 mutations.
Cell Rep, 39, 2022
|
|
8XJV
 
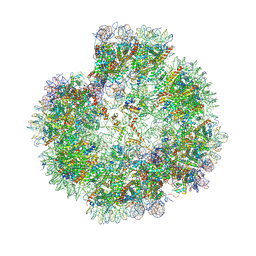 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
3KR3
 
 | | Crystal structure of IGF-II antibody complex | | Descriptor: | 1,2-ETHANEDIOL, Insulin-like growth factor II, antibody-Fab (heavy chain), ... | | Authors: | Peat, T.S, Newman, J, Adams, T.E. | | Deposit date: | 2009-11-17 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A human monoclonal antibody against insulin-like growth factor-II blocks the growth of human hepatocellular carcinoma cell lines in vitro and in vivo.
Mol.Cancer Ther., 9, 2010
|
|
5GLY
 
 | | Crystal structure of a glycoside hydrolase in complex with cellotetrose from Thielavia terrestris NRRL 8126 | | Descriptor: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
5GLX
 
 | | Crystal structure of a glycoside hydrolase from Thielavia terrestris NRRL 8126 | | Descriptor: | Glycoside hydrolase family 45 protein | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
5GM9
 
 | | Crystal structure of a glycoside hydrolase in complex with cellobiose | | Descriptor: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-13 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
7UWL
 
 | | Structure of the IL-25-IL-17RB-IL-17RA ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWM
 
 | | Structure of the IL-17A-IL-17RA binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWJ
 
 | | Structure of the homodimeric IL-25-IL-17RB binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor B, Interleukin-25 | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWK
 
 | | Structure of the higher-order IL-25-IL-17RB complex | | Descriptor: | Interleukin-17 receptor B, Interleukin-25 | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWN
 
 | | Structure of the IL-17A-IL-17RA-IL-17RC ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
6IME
 
 | | Rv2361c complex with substrate analogues | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, CARBONATE ION, ... | | Authors: | Ko, T.-P, Guo, R.-T, Chen, C.-C, Liu, W. | | Deposit date: | 2018-10-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate-analogue complex structure of Mycobacterium tuberculosis decaprenyl diphosphate synthase.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
