7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFT
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with p-hydroxybenzaldehyde | | Descriptor: | CHLORIDE ION, P-HYDROXYBENZALDEHYDE, POTASSIUM ION, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Ewert, W, Sprenger, J, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Wolf, M, Yefanov, O, Gelisio, L, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFU
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 3, 4-Dihydroxybenzoic acid, methyl ester | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Ewert, W, Werner, N, Falke, S, Guenther, S, Reinke, P, Sprenger, J, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Wolf, M, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFS
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 4-(2-hydroxyethyl)phenol | | Descriptor: | 4-(2-hydroxyethyl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Ewert, W, Sprenger, J, Koua, F, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Wolf, M, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
1JQH
 
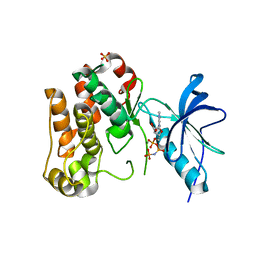 | | IGF-1 receptor kinase domain | | Descriptor: | IGF-1 receptor kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pautsch, A, Zoephel, A, Ahorn, H, Spevak, W, Hauptmann, R, Nar, H. | | Deposit date: | 2001-08-07 | | Release date: | 2002-04-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bisphosphorylated IGF-1 receptor kinase: insight into domain movements upon kinase activation.
Structure, 9, 2001
|
|
1UPV
 
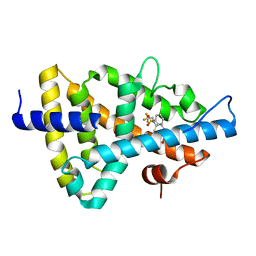 | | Crystal structure of the human Liver X receptor beta ligand binding domain in complex with a synthetic agonist | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, OXYSTEROLS RECEPTOR LXR-BETA | | Authors: | Hoerer, S, Schmid, A, Heckel, A, Budzinski, R.M, Nar, H. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human Liver X Receptor Beta Ligand-Binding Domain in Complex with a Synthetic Agonist
J.Mol.Biol., 334, 2003
|
|
1UPW
 
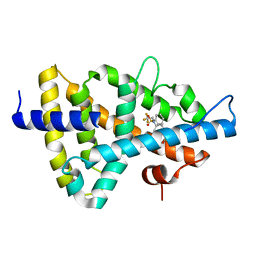 | | Crystal structure of the human Liver X receptor beta ligand binding domain in complex with a synthetic agonist | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, OXYSTEROLS RECEPTOR LXR-BETA | | Authors: | Hoerer, S, Schmid, A, Heckel, A, Budzinski, R.M, Nar, H. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human Liver X Receptor Beta Ligand-Binding Domain in Complex with a Synthetic Agonist
J.Mol.Biol., 334, 2003
|
|
1URI
 
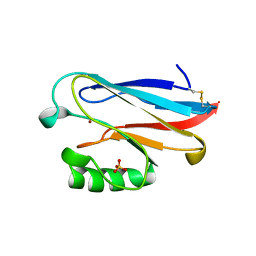 | | AZURIN MUTANT WITH MET 121 REPLACED BY GLN | | Descriptor: | AZURIN, COPPER (II) ION, SULFATE ION | | Authors: | Romero, A, Nar, H, Huber, R, Messerschmidt, A. | | Deposit date: | 1996-11-14 | | Release date: | 1997-04-01 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray analysis and spectroscopic characterization of M121Q azurin. A copper site model for stellacyanin.
J.Mol.Biol., 229, 1993
|
|
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
1ID2
 
 | | CRYSTAL STRUCTURE OF AMICYANIN FROM PARACOCCUS VERSUTUS (THIOBACILLUS VERSUTUS) | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Romero, A, Nar, H, Messerschmidt, A. | | Deposit date: | 2001-04-03 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure analysis and refinement at 2.15 A resolution of amicyanin, a type I blue copper protein, from Thiobacillus versutus.
J.Mol.Biol., 236, 1994
|
|
1AZN
 
 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT PHE114ALA FROM PSEUDOMONAS AERUGINOSA AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Pascher, T, Nar, H. | | Deposit date: | 1994-05-27 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the azurin mutant Phe114Ala from Pseudomonas aeruginosa at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
6Y0H
 
 | | High resolution structure of GH11 xylanase from Nectria haematococca | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Andaleeb, H, Betzel, C, Perbandt, M, Brognaro, H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure and biochemical characterization of a GH11 endoxylanase from Nectria haematococca.
Sci Rep, 10, 2020
|
|
8C5W
 
 | | Crystal Structure of Penicillin-binding Protein 3 (PBP3) from Staphylococcus Epidermidis in complex with Cefotaxime | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Schwinzer, M, Brognaro, H, Rohde, H, Betzel, C. | | Deposit date: | 2023-01-10 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure and Dynamics of the Penicillin-Binding Protein 3 from Staphylococcus Epidermidis Native and in Complex with Cefotaxime and Vaborbactam
Int J Appl Biol Pharm, 2023
|
|
8C5B
 
 | |
8C5O
 
 | |
1AZR
 
 | | CRYSTAL STRUCTURE OF PSEUDOMONAS AERUGINOSA ZINC AZURIN MUTANT ASP47ASP AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION, NITRATE ION | | Authors: | Sjolin, L, Tsai, Lc, Langer, V, Pascher, T, Karlsson, G, Nordling, M, Nar, H. | | Deposit date: | 1993-03-04 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Pseudomonas aeruginosai zinc azurin mutant Asn47Asp at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
4YZF
 
 | | Crystal structure of the anion exchanger domain of human erythrocyte Band 3 | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Band 3 anion transport protein, FAB fragment of Immunoglobulin (IgG) molecule | | Authors: | Alguel, Y, Arakawa, T, Yugiri, T.K, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Suno, C.I, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A.D, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-03-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the anion exchanger domain of human erythrocyte band 3.
Science, 350, 2015
|
|
5A16
 
 | | Crystal structure of Fab4201 raised against Human Erythrocyte Anion Exchanger 1 | | Descriptor: | FAB4201 HEAVY CHAIN | | Authors: | Arakawa, T, Kobayashi-Yugiri, T, Alguel, Y, Weyand, S, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Ikeda-Suno, C, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Anion Exchanger Domain of Human Erythrocyte Band 3
Science, 350, 2015
|
|
1B6Z
 
 | | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE | | Descriptor: | 6-pyruvoyl tetrahydropterin synthase, ZINC ION | | Authors: | Ploom, T, Thoeny, B, Yim, J, Lee, S, Nar, H, Leimbacher, W, Huber, R, Richardson, J, Auerbach, G. | | Deposit date: | 1999-01-18 | | Release date: | 2000-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and kinetic investigations on the mechanism of 6-pyruvoyl tetrahydropterin synthase.
J.Mol.Biol., 286, 1999
|
|
1B66
 
 | | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE | | Descriptor: | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE, BIOPTERIN, ZINC ION | | Authors: | Ploom, T, Thoeny, B, Yim, J, Lee, S, Nar, H, Leimbacher, W, Huber, R, Richardson, J, Auerbach, G. | | Deposit date: | 1999-01-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and kinetic investigations on the mechanism of 6-pyruvoyl tetrahydropterin synthase.
J.Mol.Biol., 286, 1999
|
|
7DAA
 
 | | Crystal structure of basigin complexed with anti-basigin Fab fragment | | Descriptor: | CADMIUM ION, Heavy chain of antibody Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7DCE
 
 | | Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Heavy chain of Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Tsutsumi, A, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Kikkawa, M, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D9Z
 
 | | Crystal structure of anti-basigin Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Heavy chain of antibody Fab fragment, ... | | Authors: | Sakuragi, T, Kanai, R, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.123 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5A4H
 
 | | Solution structure of the lipid droplet anchoring peptide of CGI-58 bound to DPC micelles | | Descriptor: | 1-ACYLGLYCEROL-3-PHOSPHATE O-ACYLTRANSFERASE ABHD5 | | Authors: | Boeszoermenyi, A, Arthanari, H, Wagner, G, Nagy, H.M, Zangger, K, Lindermuth, H, Oberer, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Cgi-58 Motif Provides the Molecular Basis of Lipid Droplet Anchoring.
J.Biol.Chem., 290, 2015
|
|
