8TQC
 
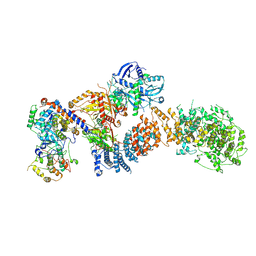 | | Structure of the human CDK8 kinase module | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, Mediator of RNA polymerase II transcription subunit 12, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-06 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 2024
|
|
8TRH
 
 | | The IDRc bound human core Mediator complex | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-09 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 2024
|
|
8TQW
 
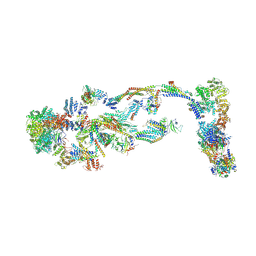 | | Structure of human transcriptional Mediator complex | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-08 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 2024
|
|
7CRD
 
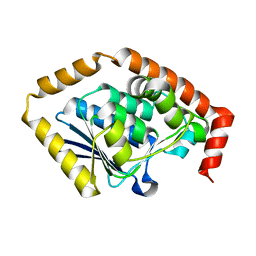 | | Structure of Pseudomonas aeruginosa OdaA | | Descriptor: | Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
8HNI
 
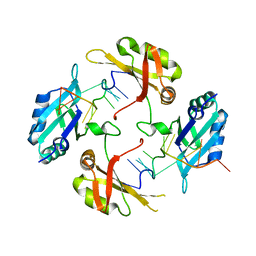 | | hnRNP A2/B1 RRMs in complex with telomeric DNA | | Descriptor: | DNA (5'-D(P*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*T)-3'), Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Liu, Y, Abula, A, Xiao, H, Guo, H, Li, T, Zheng, L, Chen, B, Nguyen, H, Ji, X. | | Deposit date: | 2022-12-07 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Structural Insight Into hnRNP A2/B1 Homodimerization and DNA Recognition.
J.Mol.Biol., 435, 2023
|
|
7CPK
 
 | | Xylanase R from Bacillus sp. TAR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endo-1,4-beta-xylanase A, ... | | Authors: | Kuwata, K, Suzuki, M, Takita, T, Nakatani, K, Li, T, Katano, Y, Kojima, K, Mizutani, K, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insight into the mechanism of thermostabilization of GH10 xylanase from Bacillus sp. strain TAR-1 by the mutation of S92 to E.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
7CPL
 
 | | Xylanase R from Bacillus sp. TAR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endo-1,4-beta-xylanase A, ... | | Authors: | Kuwata, K, Suzuki, M, Takita, T, Nakatani, K, Li, T, Katano, Y, Kojima, K, Mizutani, K, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Insight into the mechanism of thermostabilization of GH10 xylanase from Bacillus sp. strain TAR-1 by the mutation of S92 to E.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
7WM3
 
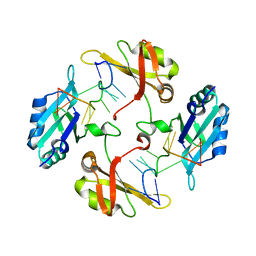 | | hnRNP A2/B1 RRMs in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(P*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Abula, A, Liu, Y, Guo, H, Li, T, Ji, X. | | Deposit date: | 2022-01-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Insight Into hnRNP A2/B1 Homodimerization and DNA Recognition.
J.Mol.Biol., 435, 2022
|
|
8IZL
 
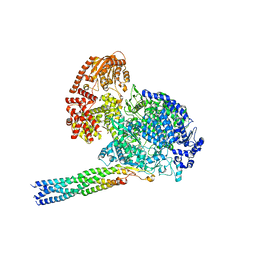 | |
9BDY
 
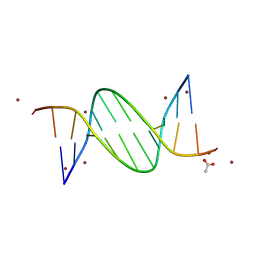 | | AT-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BE0
 
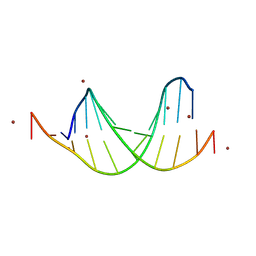 | | GC-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*GP*CP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*G)-3'), ZINC ION | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BE1
 
 | | CG-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*GP*CP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*G)-3'), ZINC ION | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Wang, V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDZ
 
 | | TA-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, ZINC ION, double-stranded DNA | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Wang, V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDX
 
 | | NF-kappaB RelA homo-dimer bound to CG-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*GP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*CP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), Transcription factor p65 | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDW
 
 | | NF-kappaB RelA homo-dimer bound to GC-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDU
 
 | | NF-kappaB RelA homo-dimer bound to AT-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDV
 
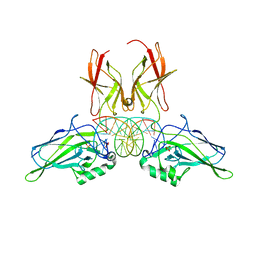 | | NF-kappaB RelA homo-dimer bound to TA-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8YXR
 
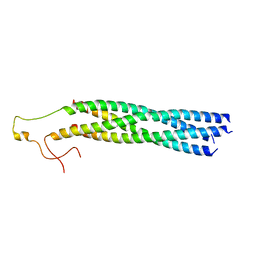 | |
8YXM
 
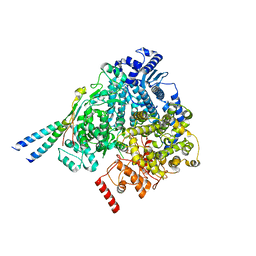 | |
8YXL
 
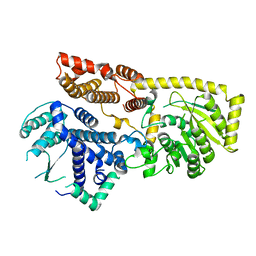 | |
8YXO
 
 | |
8YXP
 
 | | Structure of mumps virus L protein (state2) | | Descriptor: | RNA-directed RNA polymerase L, ZINC ION | | Authors: | Li, T.H, Shen, Q.T. | | Deposit date: | 2024-04-02 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures of the mumps virus polymerase complex via cryo-electron microscopy.
Nat Commun, 15, 2024
|
|
7MK7
 
 | | Augmentor domain of augmentor-beta | | Descriptor: | ALK and LTK ligand 1,Maltodextrin-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krimmer, S.G, Reshetnyak, A.V, Puleo, D.E, Schlessinger, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42815185 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
6M2Z
 
 | | Crystal structure of a formolase, BFD variant M3 from Pseudomonas putida | | Descriptor: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
6M2Y
 
 | | Crystal structure of a formolase, BFD variant M6 from Pseudomonas putida | | Descriptor: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
